Translatomics: The Global View of Translation
- PMID: 30626072
- PMCID: PMC6337585
- DOI: 10.3390/ijms20010212
Translatomics: The Global View of Translation
Abstract
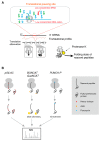
In all kingdoms of life, proteins are synthesized by ribosomes in a process referred to as translation. The amplitude of translational regulation exceeds the sum of transcription, mRNA degradation and protein degradation. Therefore, it is essential to investigate translation in a global scale. Like the other "omics"-methods, translatomics investigates the totality of the components in the translation process, including but not limited to translating mRNAs, ribosomes, tRNAs, regulatory RNAs and nascent polypeptide chains. Technical advances in recent years have brought breakthroughs in the investigation of these components at global scale, both for their composition and dynamics. These methods have been applied in a rapidly increasing number of studies to reveal multifaceted aspects of translation control. The process of translation is not restricted to the conversion of mRNA coding sequences into polypeptide chains, it also controls the composition of the proteome in a delicate and responsive way. Therefore, translatomics has extended its unique and innovative power to many fields including proteomics, cancer research, bacterial stress response, biological rhythmicity and plant biology. Rational design in translation can enhance recombinant protein production for thousands of times. This brief review summarizes the main state-of-the-art methods of translatomics, highlights recent discoveries made in this field and introduces applications of translatomics on basic biological and biomedical research.
Keywords: RNC-mRNA; RNC-seq; Ribo-seq; polysome profiling; translation regulation; translatomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Insights Into Translatomics in the Nervous System.Front Genet. 2020 Dec 21;11:599548. doi: 10.3389/fgene.2020.599548. eCollection 2020. Front Genet. 2020. PMID: 33408739 Free PMC article. Review.
-
RIVET: comprehensive graphic user interface for analysis and exploration of genome-wide translatomics data.BMC Genomics. 2018 Nov 8;19(1):809. doi: 10.1186/s12864-018-5166-z. BMC Genomics. 2018. PMID: 30409155 Free PMC article.
-
[Cis-regulatory mechanisms and biological effects of translation elongation].Yi Chuan. 2020 Jul 20;42(7):613-631. doi: 10.16288/j.yczz.20-074. Yi Chuan. 2020. PMID: 32694102 Review. Chinese.
-
GWIPS-viz as a tool for exploring ribosome profiling evidence supporting the synthesis of alternative proteoforms.Proteomics. 2015 Jul;15(14):2410-6. doi: 10.1002/pmic.201400603. Epub 2015 Apr 23. Proteomics. 2015. PMID: 25736862 Free PMC article.
-
RiboGalaxy: A browser based platform for the alignment, analysis and visualization of ribosome profiling data.RNA Biol. 2016;13(3):316-9. doi: 10.1080/15476286.2016.1141862. Epub 2016 Jan 29. RNA Biol. 2016. PMID: 26821742 Free PMC article.
Cited by
-
Workability of mRNA Sequencing for Predicting Protein Abundance.Genes (Basel). 2023 Nov 11;14(11):2065. doi: 10.3390/genes14112065. Genes (Basel). 2023. PMID: 38003008 Free PMC article. Review.
-
Insights Into Translatomics in the Nervous System.Front Genet. 2020 Dec 21;11:599548. doi: 10.3389/fgene.2020.599548. eCollection 2020. Front Genet. 2020. PMID: 33408739 Free PMC article. Review.
-
Regulating the regulator: a survey of mechanisms from transcription to translation controlling expression of mammalian cell cycle kinase Aurora A.Open Biol. 2022 Sep;12(9):220134. doi: 10.1098/rsob.220134. Epub 2022 Sep 7. Open Biol. 2022. PMID: 36067794 Free PMC article. Review.
-
Disruption of the RNA modifications that target the ribosome translation machinery in human cancer.Mol Cancer. 2020 Apr 2;19(1):70. doi: 10.1186/s12943-020-01192-8. Mol Cancer. 2020. PMID: 32241281 Free PMC article. Review.
-
A Novel Approach for Screening Sericin-Derived Therapeutic Peptides Using Transcriptomics and Immunoprecipitation.Int J Mol Sci. 2023 May 29;24(11):9425. doi: 10.3390/ijms24119425. Int J Mol Sci. 2023. PMID: 37298379 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

