Cross-Domain and Viral Interactions in the Microbiome
- PMID: 30626617
- PMCID: PMC6383444
- DOI: 10.1128/MMBR.00044-18
Cross-Domain and Viral Interactions in the Microbiome
Abstract
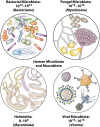
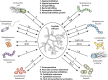
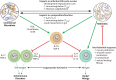
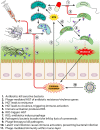
The importance of the microbiome to human health is increasingly recognized and has become a major focus of recent research. However, much of the work has focused on a few aspects, particularly the bacterial component of the microbiome, most frequently in the gastrointestinal tract. Yet humans and other animals can be colonized by a wide array of organisms spanning all domains of life, including bacteria and archaea, unicellular eukaryotes such as fungi, multicellular eukaryotes such as helminths, and viruses. As they share the same host niches, they can compete with, synergize with, and antagonize each other, with potential impacts on their host. Here, we discuss these major groups making up the human microbiome, with a focus on how they interact with each other and their multicellular host.
Keywords: archaea; bacteria; bacteriophage; cross-domain; fungi; helminths; microbiome; protozoa; virus.
Copyright © 2019 American Society for Microbiology.
Figures
References
-
- Bikel S, Valdez-Lara A, Cornejo-Granados F, Rico K, Canizales-Quinteros S, Soberón X, Del Pozo-Yauner L, Ochoa-Leyva A. 2015. Combining metagenomics, metatranscriptomics and viromics to explore novel microbial interactions: towards a systems-level understanding of human microbiome. Comput Struct Biotechnol J 13:390–401. doi: 10.1016/j.csbj.2015.06.001. - DOI - PMC - PubMed
-
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

