Selective Sweeps
- PMID: 30626638
- PMCID: PMC6325696
- DOI: 10.1534/genetics.118.301319
Selective Sweeps
Abstract
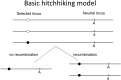
For almost 20 years, many inference methods have been developed to detect selective sweeps and localize the targets of directional selection in the genome. These methods are based on population genetic models that describe the effect of a beneficial allele (e.g., a new mutation) on linked neutral variation (driven by directional selection from a single copy to fixation). Here, I discuss these models, ranging from selective sweeps in a panmictic population of constant size to evolutionary traffic when simultaneous sweeps at multiple loci interfere, and emphasize the important role of demography and population structure in data analysis. In the past 10 years, soft sweeps that may arise after an environmental change from directional selection on standing variation have become a focus of population genetic research. In contrast to selective sweeps, they are caused by beneficial alleles that were neutrally segregating in a population before the environmental change or were present at a mutation-selection balance in appreciable frequency.
Keywords: background selection; demography; genetic hitchhiking; selective sweeps.
Copyright © 2019 by the Genetics Society of America.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

