Water-Soluble Humic Materials Regulate Quorum Sensing in Sinorhizobium meliloti Through a Novel Repressor of expR
- PMID: 30627123
- PMCID: PMC6309736
- DOI: 10.3389/fmicb.2018.03194
Water-Soluble Humic Materials Regulate Quorum Sensing in Sinorhizobium meliloti Through a Novel Repressor of expR
Abstract
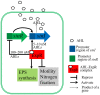
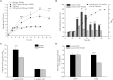
Quorum sensing (QS) plays an important role in the growth, nodulation, and nitrogen fixation of rhizobia. In this study, we show that water-soluble humic materials (WSHM) repress the expression of the QS related genes sinI, sinR, and expR in Sinorhizobium meliloti. This decreased the production of N-acetyl homoserine lactones (AHL) and exopolysaccharides (EPS), and ultimately increased S. meliloti cell density. We also identified a novel regulator, SMc03890 (renamed QsrR), which binds directly to the expR promoter. Deletion of qsrR increased expR expression. WSHM repressed the expression of expR by augmenting the interaction between QsrR and the expR promoter; this was determined by a bacterial-one-hybrid assay. These effects of WSHM on the QS system in S. meliloti may be the underlying mechanism by which WSHM increase the symbiotic nitrogen fixation of Medicago sativa inoculated with S. meliloti. This study provides the first evidence that humic acids regulate the QS of rhizobia and suggests that WSHM could be used as fertilizers to improve the efficiency of symbiotic nitrogen fixation.
Keywords: ExpR regulator; Sinorhizobium meliloti; bacterial communication; humic materials; quorum sensing.
Figures




References
-
- Bogino P. C., Nievas F. L., Giordano W. (2015). A review: quorum sensing in Bradyrhizobium. Appl. Soil Ecol. 94 49–58. 10.1016/j.apsoil.2015.04.016 - DOI
-
- Bourassa D. V., Kannenberg E. L., Sherrier D. J., Buhr R. J., Carlson R. W. (2017). The lipopolysaccharide lipid a long-chain fatty acid is important for Rhizobium leguminosarum growth and stress adaptation in free-living and nodule environments. Mol. Plant Microbe Interact. 30 161–175. 10.1094/MPMI-11-16-0230-R - DOI - PubMed
-
- Canellas L. P., Olivares F. L., Aguiar N. O., Jones D. L., Nebbioso A., Mazzei P., et al. (2015). Humic and fulvic acids as biostimulants in horticulture. Sci. Hortic. 196 15–27. 10.1016/j.scienta.2015.09.013 - DOI
LinkOut - more resources
Full Text Sources

