Spectroscopic and Electrochemical Characterization of the Mycofactocin Biosynthetic Protein, MftC, Provides Insight into Its Redox Flipping Mechanism
- PMID: 30628436
- PMCID: PMC6380935
- DOI: 10.1021/acs.biochem.8b01082
Spectroscopic and Electrochemical Characterization of the Mycofactocin Biosynthetic Protein, MftC, Provides Insight into Its Redox Flipping Mechanism
Abstract
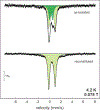
Mycofactocin is a putative redox cofactor and is classified as a ribosomally synthesized and post-translationally modified peptide (RiPP). Some RiPP natural products, including mycofactocin, rely on a radical S-adenosylmethionine (RS, SAM) protein to modify the precursor peptide. Mycofactocin maturase, MftC, is a unique RS protein that catalyzes the oxidative decarboxylation and C-C bond formation on the precursor peptide MftA. However, the number, chemical nature, and catalytic roles for the MftC [Fe-S] clusters remain unknown. Here, we report that MftC binds a RS [4Fe-4S] cluster and two auxiliary [4Fe-4S] clusters that are required for MftA modification. Furthermore, electron paramagnetic resonance spectra of MftC suggest that SAM and MftA affect the environments of the RS and Aux I cluster, whereas the Aux II cluster is unaffected by the substrates. Lastly, reduction potential assignments of individual [4Fe-4S] clusters by protein film voltammetry show that their potentials are within 100 mV of each other.
Figures
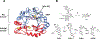
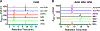
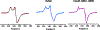



References
-
- Barr I, Latham JA, Iavarone AT, Chantarojsiri T, Hwang JD, and Klinman JP (2016) The pyrroloquinoline quinone (PQQ) biosynthetic pathway: Demonstration of de novo carbon-carbon cross-linking within the peptide substrate (PqqA) in the presence of the radical SAM enzyme (PqqE) and its peptide chaperone (PqqD). J. Biol. Chem 291, 8877–8884. - PMC - PubMed
-
- Flühe L, Knappe T. a, Gattner MJ, Schäfer A, Burghaus O, Linne U, and Marahiel M. a. (2012) The radical SAM enzyme AlbA catalyzes thioether bond formation in subtilosin A. Nat. Chem. Biol 8, 350–357. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

