A thirteen‑gene set efficiently predicts the prognosis of glioblastoma
- PMID: 30628650
- PMCID: PMC6390043
- DOI: 10.3892/mmr.2019.9801
A thirteen‑gene set efficiently predicts the prognosis of glioblastoma
Abstract
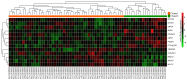
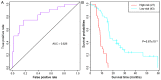
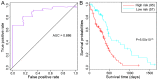
Glioblastoma multiforme (GBM) is the most common type of brain cancer; it usually recurs and patients have a short survival time. The present study aimed to construct a gene expression classifier and to screen key genes associated with GBM prognosis. GSE7696 microarray data set included samples from 10 recurrent GBM tissues, 70 primary GBM tissues and 4 normal brain tissues. Seed genes were identified by the 'survival' package in R and subjected to pathway enrichment analysis. Prognostic genes were selected from the seed genes using the 'rbsurv' package in R, unsupervised hierarchical clustering, survival analysis and enrichment analysis. Multivariate survival analysis was performed for the prognostic genes, and the GBM data set from The Cancer Genome Atlas database was utilized to validate the prognostic genes. Of the 1,785 seed genes analyzed, 13 prognostic feature genes, including collagen type XXVIII α1 chain (COL28A1), PDS5 cohesin‑associated factor A (PDS5A), zinc‑finger DHHC‑type containing 2 (ZDHHC2), zinc‑finger protein 24 (ZNF24), myosin VA (MYO5A) and myeloid/lymphoid or mixed‑lineage leukemia translocated to 4 (MLLT4), were identified. These genes performed well on sample classification and prognostic risk differentiation, and six pathways, including adherens junction, cyclic adenosine 3',5'‑monophosphate signaling and Ras signaling pathways, were enriched for these feature genes. The high‑risk group was slightly older compared with the low‑risk group. The validation data set confirmed the prognostic value of the 13 feature genes for GBM; of these, COL28A1, PDS5A, ZDHHC2, ZNF24, MYO5A and MLLT4 may be crucial. These results may aid the understanding of the pathogenesis of GBM and provide important clues for the development of novel diagnostic markers or therapeutic targets.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

