In vitro activity of apramycin against multidrug-, carbapenem- and aminoglycoside-resistant Enterobacteriaceae and Acinetobacter baumannii
- PMID: 30629184
- PMCID: PMC6419615
- DOI: 10.1093/jac/dky546
In vitro activity of apramycin against multidrug-, carbapenem- and aminoglycoside-resistant Enterobacteriaceae and Acinetobacter baumannii
Abstract
Objectives: Widespread antimicrobial resistance often limits the availability of therapeutic options to only a few last-resort drugs that are themselves challenged by emerging resistance and adverse side effects. Apramycin, an aminoglycoside antibiotic, has a unique chemical structure that evades almost all resistance mechanisms including the RNA methyltransferases frequently encountered in carbapenemase-producing clinical isolates. This study evaluates the in vitro activity of apramycin against multidrug-, carbapenem- and aminoglycoside-resistant Enterobacteriaceae and Acinetobacter baumannii, and provides a rationale for its superior antibacterial activity in the presence of aminoglycoside resistance determinants.
Methods: A thorough antibacterial assessment of apramycin with 1232 clinical isolates from Europe, Asia, Africa and South America was performed by standard CLSI broth microdilution testing. WGS and susceptibility testing with an engineered panel of aminoglycoside resistance-conferring determinants were used to provide a mechanistic rationale for the breadth of apramycin activity.
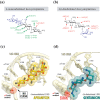
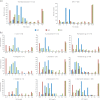
Results: MIC distributions and MIC90 values demonstrated broad antibacterial activity of apramycin against Escherichia coli, Klebsiella pneumoniae, Enterobacter spp., Morganella morganii, Citrobacter freundii, Providencia spp., Proteus mirabilis, Serratia marcescens and A. baumannii. Genotypic analysis revealed the variety of aminoglycoside-modifying enzymes and rRNA methyltransferases that rendered a remarkable proportion of clinical isolates resistant to standard-of-care aminoglycosides, but not to apramycin. Screening a panel of engineered strains each with a single well-defined resistance mechanism further demonstrated a lack of cross-resistance to gentamicin, amikacin, tobramycin and plazomicin.
Conclusions: Its superior breadth of activity renders apramycin a promising drug candidate for the treatment of systemic Gram-negative infections that are resistant to treatment with other aminoglycoside antibiotics.
© The Author(s) 2019. Published by Oxford University Press on behalf of the British Society for Antimicrobial Chemotherapy.
Figures
References
-
- Potron A, Poirel L, Nordmann P.. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int J Antimicrob Agents 2015; 45: 568–85. - PubMed
-
- Busani S, Serafini G, Mantovani E. et al. Mortality in patients with septic shock by multidrug resistant bacteria. J Intensive Care Med 2017; doi: 10.1177/0885066616688165. - PubMed
-
- Poirel L, Kieffer N, Liassine N. et al. Plasmid-mediated carbapenem and colistin resistance in a clinical isolate of Escherichia coli. Lancet Infect Dis 2016; 16: 281. - PubMed
-
- WHO. Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-resistant Bacterial Infections. Geneva: World Health Organization, 2017. (WHO/EMP/IAU/2017.12).
-
- Blair JM, Webber MA, Baylay AJ. et al. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol 2015; 13: 42–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

