Phylogeographic analysis of human influenza A and B viruses in Myanmar, 2010-2015
- PMID: 30629691
- PMCID: PMC6328249
- DOI: 10.1371/journal.pone.0210550
Phylogeographic analysis of human influenza A and B viruses in Myanmar, 2010-2015
Abstract
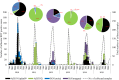
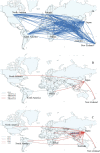
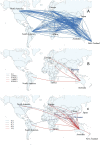
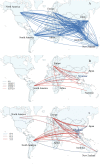
We investigated the circulation patterns of human influenza A and B viruses in Myanmar between 2010 and 2015 by analyzing full HA genes. Upper respiratory tract specimens were collected from patients with symptoms of influenza-like illness. A total of 2,860 respiratory samples were screened by influenza rapid diagnostic test, of which 1,577 (55.1%) and 810 (28.3%) were positive for influenza A and B, respectively. Of the 1,010 specimens that were positive for virus isolation, 370 (36.6%) were A(H1N1)pdm09, 327 (32.4%) were A(H3N2), 130 (12.9%) B(Victoria), and 183 (18.1%) were B(Yamagata) viruses. Our data showed that influenza epidemics mainly occurred during the rainy season in Myanmar. Our three study sites, Yangon, Pyinmana, and Pyin Oo Lwin had similar seasonality and circulating type and subtype of influenza in a given year. Moreover, viruses circulating in Myanmar during the study period were closely related genetically to those detected in Thailand, India, and China. Phylogeographic analysis showed that A(H1N1)pdm09 viruses in Myanmar originated from Europe and migrated to other countries via Japan. Similarly, A(H3N2) viruses in Myanmar originated from Europe, and disseminated to the various countries via Australia. In addition, Myanmar plays a key role in reseeding of influenza B viruses to Southeast Asia and East Asia as well as Europe and Africa. Thus, we concluded that influenza virus in Myanmar has a strong link to neighboring Asian countries, Europe and Oceania.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Cox N, Subbarao K. Global epidemiology of influenza: past and present. Annual review of medicine. 2000;51(1):407–21. - PubMed
-
- Organization WH. Manual for the laboratory diagnosis and virological surveillance of influenza. 2011.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

