HIV-1 Neutralizing Antibody Signatures and Application to Epitope-Targeted Vaccine Design
- PMID: 30629920
- PMCID: PMC6331341
- DOI: 10.1016/j.chom.2018.12.001
HIV-1 Neutralizing Antibody Signatures and Application to Epitope-Targeted Vaccine Design
Erratum in
-
HIV-1 Neutralizing Antibody Signatures and Application to Epitope-Targeted Vaccine Design.Cell Host Microbe. 2019 Aug 14;26(2):296. doi: 10.1016/j.chom.2019.07.016. Cell Host Microbe. 2019. PMID: 31415756 Free PMC article. No abstract available.
Abstract
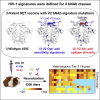
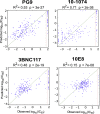
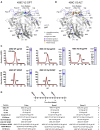
Eliciting HIV-1-specific broadly neutralizing antibodies (bNAbs) remains a challenge for vaccine development, and the potential of passively delivered bNAbs for prophylaxis and therapeutics is being explored. We used neutralization data from four large virus panels to comprehensively map viral signatures associated with bNAb sensitivity, including amino acids, hypervariable region characteristics, and clade effects across four different classes of bNAbs. The bNAb signatures defined for the variable loop 2 (V2) epitope region of HIV-1 Env were then employed to inform immunogen design in a proof-of-concept exploration of signature-based epitope targeted (SET) vaccines. V2 bNAb signature-guided mutations were introduced into Env 459C to create a trivalent vaccine, and immunization of guinea pigs with V2-SET vaccines resulted in increased breadth of NAb responses compared with Env 459C alone. These data demonstrate that bNAb signatures can be utilized to engineer HIV-1 Env vaccine immunogens capable of eliciting antibody responses with greater neutralization breadth.
Keywords: HIV-1; V2-apex antibodies; broadly neutralizing antibodies; hypervariable regions; machine learning; signature analysis; vaccine design.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Arlot S., Celisse A. A survey of cross-validation procedures for model selection. Statist. Surv. 2010;4:40–79.
-
- Bhattacharya T., Daniels M., Heckerman D., Foley B., Frahm N., Kadie C., Carlson J., Yusim K., McMahon B., Gaschen B. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science. 2007;315:1583–1586. - PubMed
-
- Bonsignori M., Hwang K.K., Chen X., Tsao C.Y., Morris L., Gray E., Marshall D.J., Crump J.A., Kapiga S.H., Sam N.E. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J. Virol. 2011;85:9998–10009. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

