The N-cadherin interactome in primary cardiomyocytes as defined using quantitative proximity proteomics
- PMID: 30630894
- PMCID: PMC6382013
- DOI: 10.1242/jcs.221606
The N-cadherin interactome in primary cardiomyocytes as defined using quantitative proximity proteomics
Abstract
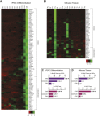
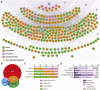
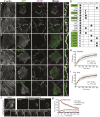
The junctional complexes that couple cardiomyocytes must transmit the mechanical forces of contraction while maintaining adhesive homeostasis. The adherens junction (AJ) connects the actomyosin networks of neighboring cardiomyocytes and is required for proper heart function. Yet little is known about the molecular composition of the cardiomyocyte AJ or how it is organized to function under mechanical load. Here, we define the architecture, dynamics and proteome of the cardiomyocyte AJ. Mouse neonatal cardiomyocytes assemble stable AJs along intercellular contacts with organizational and structural hallmarks similar to mature contacts. We combine quantitative mass spectrometry with proximity labeling to identify the N-cadherin (CDH2) interactome. We define over 350 proteins in this interactome, nearly 200 of which are unique to CDH2 and not part of the E-cadherin (CDH1) interactome. CDH2-specific interactors comprise primarily adaptor and adhesion proteins that promote junction specialization. Our results provide novel insight into the cardiomyocyte AJ and offer a proteomic atlas for defining the molecular complexes that regulate cardiomyocyte intercellular adhesion. This article has an associated First Person interview with the first authors of the paper.
Keywords: Adherens junction; CDH2; Cardiomyocyte; Interactome; Intercalated disc; N-cadherin; Proteomics.
© 2019. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures




References
-
- Angst B. D., Khan L. U. R., Severs N. J., Whitely K., Rothery S., Thompson R. P., Magee A. I. and Gourdie R. G. (1997). Dissociated spatial patterning of gap junctions and cell adhesion junctions during postnatal differentiation of ventricular myocardium. Circ. Res. 80, 88-94. 10.1161/01.RES.80.1.88 - DOI - PubMed
-
- Borrmann C. M., Grund C., Kuhn C., Hofmann I., Pieperhoff S. and Franke W. W. (2006). The area composita of adhering junctions connecting heart muscle cells of vertebrates. II. Colocalizations of desmosomal and fascia adhaerens molecules in the intercalated disk. Eur. J. Cell Biol. 85, 469-485. 10.1016/j.ejcb.2006.02.009 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

