Nucleosome spacing periodically modulates nucleosome chain folding and DNA topology in circular nucleosome arrays
- PMID: 30630950
- PMCID: PMC6422092
- DOI: 10.1074/jbc.RA118.006412
Nucleosome spacing periodically modulates nucleosome chain folding and DNA topology in circular nucleosome arrays
Abstract
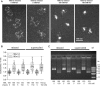
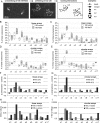
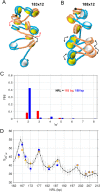
The length of linker DNA that separates nucleosomes is highly variable, but its mechanistic role in modulating chromatin structure and functions remains unknown. Here, we established an experimental system using circular arrays of positioned nucleosomes to investigate whether variations in nucleosome linker length could affect nucleosome folding, self-association, and interactions. We conducted EM, DNA topology, native electrophoretic assays, and Mg2+-dependent self-association assays to study intrinsic folding of linear and circular nucleosome arrays with linker DNA length of 36 bp and 41 bp (3.5 turns and 4 turns of DNA double helix, respectively). These experiments revealed that potential artifacts arising from open DNA ends and full DNA relaxation in the linear arrays do not significantly affect overall chromatin compaction and self-association. We observed that the 0.5 DNA helical turn difference between the two DNA linker lengths significantly affects DNA topology and nucleosome interactions. In particular, the 41-bp linkers promoted interactions between any two nucleosome beads separated by one bead as expected for a zigzag fiber, whereas the 36-bp linkers promoted interactions between two nucleosome beads separated by two other beads and also reduced negative superhelicity. Monte Carlo simulations accurately reproduce periodic modulations of chromatin compaction, DNA topology, and internucleosomal interactions with a 10-bp periodicity. We propose that the nucleosome spacing and associated chromatin structure modulations may play an important role in formation of different chromatin epigenetic states, thus suggesting implications for how chromatin accessibility to DNA-binding factors and the RNA transcription machinery is regulated.
Keywords: DNA topology; chromatin higher order structure; chromatin structure; computer modeling; conformational simulation; electron microscopy (EM); epigenetic regulation; epigenetics; gene regulation; histone; linker DNA; nucleosome.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

