Characterizing both bacteria and fungi improves understanding of the Arabidopsis root microbiome
- PMID: 30631088
- PMCID: PMC6328596
- DOI: 10.1038/s41598-018-37208-z
Characterizing both bacteria and fungi improves understanding of the Arabidopsis root microbiome
Abstract
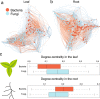
Roots provide plants mineral nutrients and stability in soil; while doing so, they come into contact with diverse soil microbes that affect plant health and productivity. Despite their ecological and agricultural relevance, the factors that shape the root microbiome remain poorly understood. We grew a worldwide panel of replicated Arabidopsis thaliana accessions outdoors and over winter to characterize their root-microbial communities. Although studies of the root microbiome tend to focus on bacteria, we found evidence that fungi have a strong influence on the structure of the root microbiome. Moreover, host effects appear to have a stronger influence on plant-fungal communities than plant-bacterial communities. Mapping the host genes that affect microbiome traits identified a priori candidate genes with roles in plant immunity; the root microbiome also appears to be strongly affected by genes that impact root and root hair development. Our results suggest that future analyses of the root microbiome should focus on multiple kingdoms, and that the root microbiome is shaped not only by genes involved in defense, but also by genes involved in plant form and physiology.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

