Large scale comparison of QSAR and conformal prediction methods and their applications in drug discovery
- PMID: 30631996
- PMCID: PMC6690068
- DOI: 10.1186/s13321-018-0325-4
Large scale comparison of QSAR and conformal prediction methods and their applications in drug discovery
Abstract
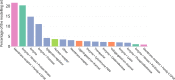
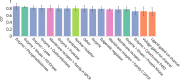
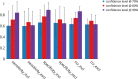
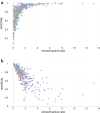
Structure-activity relationship modelling is frequently used in the early stage of drug discovery to assess the activity of a compound on one or several targets, and can also be used to assess the interaction of compounds with liability targets. QSAR models have been used for these and related applications over many years, with good success. Conformal prediction is a relatively new QSAR approach that provides information on the certainty of a prediction, and so helps in decision-making. However, it is not always clear how best to make use of this additional information. In this article, we describe a case study that directly compares conformal prediction with traditional QSAR methods for large-scale predictions of target-ligand binding. The ChEMBL database was used to extract a data set comprising data from 550 human protein targets with different bioactivity profiles. For each target, a QSAR model and a conformal predictor were trained and their results compared. The models were then evaluated on new data published since the original models were built to simulate a "real world" application. The comparative study highlights the similarities between the two techniques but also some differences that it is important to bear in mind when the methods are used in practical drug discovery applications.
Keywords: ChEMBL; Cheminformatics; Classification models; Mondrian conformal prediction; QSAR.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Verma J, Khedkar V, Coutinho E. 3D-QSAR in drug design: a review. Curr Top Med Chem. 2010;10:95–115. - PubMed
-
- Quintero FA, Patel SJ, Muñoz F, Sam Mannan M. Review of existing QSAR/QSPR models developed for properties used in hazardous chemicals classification system. Ind Eng Chem Res. 2012;51:16101–16115.
Grants and funding
LinkOut - more resources
Full Text Sources

