Matrix metalloprotease-1 inhibits and disrupts Enterococcus faecalis biofilms
- PMID: 30633757
- PMCID: PMC6329490
- DOI: 10.1371/journal.pone.0210218
Matrix metalloprotease-1 inhibits and disrupts Enterococcus faecalis biofilms
Abstract
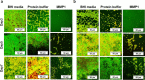
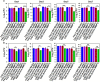
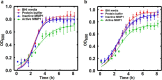
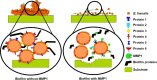
Enterococcus faecalis is a major opportunistic pathogen that readily forms protective biofilms leading to chronic infections. Biofilms protect bacteria from detergent solutions, antimicrobial agents, environmental stress, and effectively make bacteria 10 to 1000-fold more resistant to antibiotic treatment. Extracellular proteins and polysaccharides are primary components of biofilms and play a key role in cell survival, microbial persistence, cellular interaction, and maturation of E. faecalis biofilms. Degradation of biofilm components by mammalian proteases is an effective antibiofilm strategy because proteases are known to degrade bacterial proteins leading to bacterial cell lysis and growth inhibition. Here, we show that human matrix metalloprotease-1 inhibits and disrupts E. faecalis biofilms. MMPs are cell-secreted zinc- and calcium-dependent proteases that degrade and regulate various structural components of the extracellular matrix. Human MMP1 is known to degrade type-1 collagen and can also cleave a wide range of substrates. We found that recombinant human MMP1 significantly inhibited and disrupted biofilms of vancomycin sensitive and vancomycin resistant E. faecalis strains. The mechanism of antibiofilm activity is speculated to be linked with bacterial growth inhibition and degradation of biofilm matrix proteins by MMP1. These findings suggest that human MMP1 can potentially be used as a potent antibiofilm agent against E. faecalis biofilms.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Control CfD, Prevention. Antibiotic resistance threats in the United States, 2013: Centres for Disease Control and Prevention, US Department of Health and Human Services; 2013.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

