Analysis of the Bacterial and Host Proteins along and across the Porcine Gastrointestinal Tract
- PMID: 30634649
- PMCID: PMC6473940
- DOI: 10.3390/proteomes7010004
Analysis of the Bacterial and Host Proteins along and across the Porcine Gastrointestinal Tract
Abstract
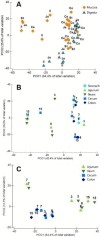
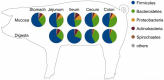
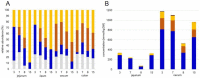
Pigs are among the most important farm animals worldwide and research to optimize their feed efficiency and improve their welfare is still in progress. The porcine intestinal microbiome is so far mainly known from sequencing-based studies. Digesta and mucosa samples from five different porcine gastrointestinal tract sections were analyzed by metaproteomics to obtain a deeper insight into the functions of bacterial groups with concomitant analyses of host proteins. Firmicutes (Prevotellaceae) dominated mucosa and digesta samples, followed by Bacteroidetes. Actinobacteria and Proteobacteria were much higher in abundance in mucosa compared to digesta samples. Functional profiling reveals the presence of core functions shared between digesta and mucosa samples. Protein abundances of energy production and conversion were higher in mucosa samples, whereas in digesta samples more proteins were involved in lipid transport and metabolism; short-chain fatty acids production were detected. Differences were also highlighted between sections, with the small intestine appearing more involved in carbohydrate transport and metabolism than the large intestine. Thus, this study produced the first functional analyses of the porcine GIT biology, discussing the findings in relation to expected bacterial and host functions.
Keywords: metaproteomics; microbiome; pig.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Hill J.E., Hemmingsen S.M., Goldade B.G., Dumonceaux T.J., Klassen J., Zijlstra R.T., Goh S.H., Van Kessel A.G. Comparison of ileum microflora of pigs fed corn-, wheat-, or barley-based diets by chaperonin-60 sequencing and quantitative PCR. Appl. Environ. Microbiol. 2005;71:867–875. doi: 10.1128/AEM.71.2.867-875.2005. - DOI - PMC - PubMed
-
- Richards J., Gong J., De Lange C. The gastrointestinal microbiota and its role in monogastric nutrition and health with an emphasis on pigs: Current understanding, possible modulations, and new technologies for ecological studies. Can. J. Anim Sci. 2005;85:421–435. doi: 10.4141/A05-049. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

