Single cell sequencing reveals gene expression signatures associated with bone marrow stromal cell subpopulations and time in culture
- PMID: 30635013
- PMCID: PMC6330466
- DOI: 10.1186/s12967-018-1766-2
Single cell sequencing reveals gene expression signatures associated with bone marrow stromal cell subpopulations and time in culture
Abstract
Background: Bone marrow stromal cells (BMSCs) are a heterogeneous population that participates in wound healing, immune modulation and tissue regeneration. Next generation sequencing was used to analyze transcripts from single BMSCs in order to better characterize BMSC subpopulations.
Methods: Cryopreserved passage 2 BMSCs from one healthy subject were cultured through passage 10. The transcriptomes of bulk BMSCs from designated passages were analyzed with microarrays and RNA sequencing (RNA-Seq). For some passages, single BMSCs were separated using microfluidics and their transcriptomes were analyzed by RNA-Seq.
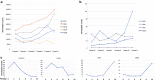
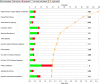
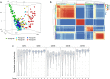
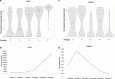
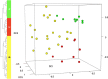
Results: Transcriptome analysis by microarray and RNA-Seq of unseparated BMSCs from passages 2, 4, 6, 8, 9 and 10 yielded similar results; both data sets grouped passages 4 and 6 and passages 9 and 10 together and genes differentially expressed among these early and late passage BMSCs were similar. 3D Diffusion map visualization of single BMSCs from passages 3, 4, 6, 8 and 9 clustered passages 3 and 9 into two distinct groups, but there was considerable overlap for passages 4, 6 and 8 cells. Markers for early passage, FGFR2, and late passage BMSCs, PLAT, were able to identify three subpopulations within passage 3 BMSCs; one that expressed high levels of FGFR2 and low levels of PLAT; one that expressed low levels of FGFR2 and high levels of PLAT and one that expressed intermediate levels of FGFR2 and low levels of PLAT.
Conclusions: Single BMSCs can be separated by microfluidics and their transcriptome analyzed by next generation sequencing. Single cell analysis of early passage BMSCs identified a subpopulation of cells expressing high levels of FGFR2 that might include skeletal stem cells.
Keywords: Bone marrow stromal cells; FGFR2; Next generation sequencing; PLAT; Single cell next generation sequencing.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

