Two Novel α-l-Arabinofuranosidases from Bifidobacterium longum subsp. longum Belonging to Glycoside Hydrolase Family 43 Cooperatively Degrade Arabinan
- PMID: 30635377
- PMCID: PMC6414367
- DOI: 10.1128/AEM.02582-18
Two Novel α-l-Arabinofuranosidases from Bifidobacterium longum subsp. longum Belonging to Glycoside Hydrolase Family 43 Cooperatively Degrade Arabinan
Abstract
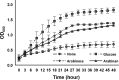
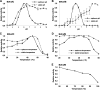
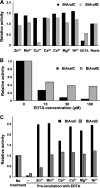
Arabinose-containing poly- or oligosaccharides are suitable carbohydrate sources for Bifidobacterium longum subsp. longum However, their degradation pathways are poorly understood. In this study, we cloned and characterized the previously uncharacterized glycoside hydrolase family 43 (GH43) enzymes B. longum subsp. longum ArafC (BlArafC; encoded by BLLJ_1852) and B. longum subsp. longum ArafB (BlArafB; encoded by BLLJ_1853) from B. longum subsp. longum JCM 1217. Both enzymes exhibited α-l-arabinofuranosidase activity toward p-nitrophenyl-α-l-arabinofuranoside but no activity toward p-nitrophenyl-β-d-xylopyranoside. The specificities of the two enzymes for l-arabinofuranosyl linkages were different. BlArafC catalyzed the hydrolysis of α1,2- and α1,3-l-arabinofuranosyl linkages found on the side chains of both arabinan and arabinoxylan. It released l-arabinose 100 times faster from arabinan than from arabinoxylan but did not act on arabinogalactan. On the other hand, BlArafB catalyzed the hydrolysis of the α1,5-l-arabinofuranosyl linkage found on the arabinan backbone. It released l-arabinose from arabinan but not from arabinoxylan or arabinogalactan. Coincubation of BlArafC and BlArafB revealed that these two enzymes are able to degrade arabinan in a synergistic manner. Both enzyme activities were suppressed with EDTA treatment, suggesting that they require divalent metal ions. The GH43 domains of BlArafC and BlArafB are classified into GH43 subfamilies 27 and 22, respectively, but show very low similarity (less than 15% identity) with other biochemically characterized members in the corresponding subfamilies. The B. longum subsp. longum strain lacking the GH43 gene cluster that includes BLLJ_1850 to BLLJ_1853 did not grow in arabinan medium, suggesting that BlArafC and BlArafB are important for assimilation of arabinan.IMPORTANCE We identified two novel α-l-arabinofuranosidases, BlArafC and BlArafB, from B. longum subsp. longum JCM 1217, both of which are predicted to be extracellular membrane-bound enzymes. The former specifically acts on α1,2/3-l-arabinofuranosyl linkages, while the latter acts on the α1,5-l-arabinofuranosyl linkage. These enzymes cooperatively degrade arabinan and are required for the efficient growth of bifidobacteria in arabinan-containing medium. The genes encoding these enzymes are located side by side in a gene cluster involved in metabolic pathways for plant-derived polysaccharides, which may confer adaptability in adult intestines.
Keywords: dietary fiber; gut microbiota; hemicellulose; intestinal microbiota; prebiotics; probiotics.
Copyright © 2019 American Society for Microbiology.
Figures



References
-
- Rogowski A, Briggs JA, Mortimer JC, Tryfona T, Terrapon N, Lowe EC, Baslé A, Morland C, Day AM, Zheng H, Rogers TE, Thompson P, Hawkins AR, Yadav MP, Henrissat B, Martens EC, Dupree P, Gilbert HJ, Bolam DN. 2015. Glycan complexity dictates microbial resource allocation in the large intestine. Nat Commun 6:7481. doi: 10.1038/ncomms8481. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

