Molecular and Functional Analysis of the Type IV Pilus Gene Cluster in Streptococcus sanguinis SK36
- PMID: 30635384
- PMCID: PMC6414370
- DOI: 10.1128/AEM.02788-18
Molecular and Functional Analysis of the Type IV Pilus Gene Cluster in Streptococcus sanguinis SK36
Abstract
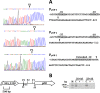
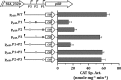
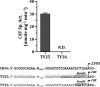
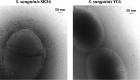
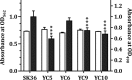
Streptococcus sanguinis, dominant in the oral microbiome, is the only known streptococcal species possessing a pil gene cluster for the biosynthesis of type IV pili (Tfp). Although this cluster is commonly present in the genome of S. sanguinis, most of the strains do not express Tfp-mediated twitching motility. Thus, this study was designed to investigate the biological functions encoded by the cluster in the twitching-negative strain S. sanguinis SK36. We found that the cluster was transcribed as an operon, with three promoters located 5' to the cluster and one in the intergenic region between SSA_2307 and SSA_2305. Studies using promoter-cat fusion strains revealed that the transcription of the cluster was mainly driven by the distal 5' promoter, which is located more than 800 bases 5' to the first gene of the cluster, SSA_2318. Optimal expression of the cluster occurred at the early stationary growth phase in a CcpA-dependent manner, although a CcpA-binding consensus is absent in the promoter region. Expression of the cluster resulted in a short hairlike surface structure under transmission electron microscopy. Deletion of the putative pilin genes (SSA_2313 to SSA_2315) abolished the biosynthesis of this structure and significantly reduced the adherence of SK36 to HeLa and SCC-4 cells. Mutations in the pil genes downregulated biofilm formation by S. sanguinis SK36. Taken together, the results demonstrate that Tfp of SK36 are important for host cell adherence, but not for motility, and that expression of the pil cluster is subject to complex regulation.IMPORTANCE The proteins and assembly machinery of the type IV pili (Tfp) are conserved throughout bacteria and archaea, and yet the function of this surface structure differs from species to species and even from strain to strain. As seen in Streptococcus sanguinis SK36, the expression of the Tfp gene cluster results in a hairlike surface structure that is much shorter than the typical Tfp. This pilus is essential for the adherence of SK36 but is not involved in motility. Being a member of the highly diverse dental biofilm, perhaps S. sanguinis could more effectively utilize this structure to adhere to host cells and to interact with other microbes within the same niche.
Keywords: CcpA; Streptococcus sanguinis; adherence; biofilms; type IV pilus.
Copyright © 2019 American Society for Microbiology.
Figures

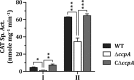

Similar articles
-
Monoderm bacteria: the new frontier for type IV pilus biology.Mol Microbiol. 2019 Dec;112(6):1674-1683. doi: 10.1111/mmi.14397. Epub 2019 Oct 8. Mol Microbiol. 2019. PMID: 31556183 Free PMC article. Review.
-
Functional Analysis of the Major Pilin Proteins of Type IV Pili in Streptococcus sanguinis CGMH010.Int J Mol Sci. 2024 May 15;25(10):5402. doi: 10.3390/ijms25105402. Int J Mol Sci. 2024. PMID: 38791440 Free PMC article.
-
Prevalence of Type IV Pili-Mediated Twitching Motility in Streptococcus sanguinis Strains and Its Impact on Biofilm Formation and Host Adherence.Appl Environ Microbiol. 2022 Sep 22;88(18):e0140322. doi: 10.1128/aem.01403-22. Epub 2022 Sep 12. Appl Environ Microbiol. 2022. PMID: 36094177 Free PMC article.
-
Type IV Pili of Streptococcus sanguinis Contribute to Pathogenesis in Experimental Infective Endocarditis.Microbiol Spectr. 2021 Dec 22;9(3):e0175221. doi: 10.1128/Spectrum.01752-21. Epub 2021 Nov 10. Microbiol Spectr. 2021. PMID: 34756087 Free PMC article.
-
Type II secretion and type IV pili of Francisella.Ann N Y Acad Sci. 2007 Jun;1105:187-201. doi: 10.1196/annals.1409.016. Epub 2007 Apr 13. Ann N Y Acad Sci. 2007. PMID: 17435117 Review.
Cited by
-
Manganese Depletion Leads to Multisystem Changes in the Transcriptome of the Opportunistic Pathogen Streptococcus sanguinis.Front Microbiol. 2020 Nov 5;11:592615. doi: 10.3389/fmicb.2020.592615. eCollection 2020. Front Microbiol. 2020. PMID: 33250881 Free PMC article.
-
Monoderm bacteria: the new frontier for type IV pilus biology.Mol Microbiol. 2019 Dec;112(6):1674-1683. doi: 10.1111/mmi.14397. Epub 2019 Oct 8. Mol Microbiol. 2019. PMID: 31556183 Free PMC article. Review.
-
Bacterial biofilms in the human body: prevalence and impacts on health and disease.Front Cell Infect Microbiol. 2023 Aug 30;13:1237164. doi: 10.3389/fcimb.2023.1237164. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37712058 Free PMC article. Review.
-
Functional Analysis of the Major Pilin Proteins of Type IV Pili in Streptococcus sanguinis CGMH010.Int J Mol Sci. 2024 May 15;25(10):5402. doi: 10.3390/ijms25105402. Int J Mol Sci. 2024. PMID: 38791440 Free PMC article.
-
Transcriptome, Phenotypic, and Virulence Analysis of Streptococcus sanguinis SK36 Wild Type and Its CcpA-Null Derivative (ΔCcpA).Front Cell Infect Microbiol. 2019 Dec 4;9:411. doi: 10.3389/fcimb.2019.00411. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31867286 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

