Sequential evolution of virulence and resistance during clonal spread of community-acquired methicillin-resistant Staphylococcus aureus
- PMID: 30635416
- PMCID: PMC6358666
- DOI: 10.1073/pnas.1814265116
Sequential evolution of virulence and resistance during clonal spread of community-acquired methicillin-resistant Staphylococcus aureus
Erratum in
-
Correction for Copin et al., Sequential evolution of virulence and resistance during clonal spread of community-acquired methicillin-resistant Staphylococcus aureus.Proc Natl Acad Sci U S A. 2019 Mar 5;116(10):4747. doi: 10.1073/pnas.1902207116. Epub 2019 Feb 25. Proc Natl Acad Sci U S A. 2019. PMID: 30804198 Free PMC article. No abstract available.
Abstract
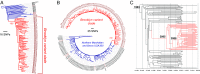
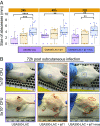
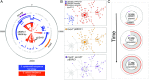
The past two decades have witnessed an alarming expansion of staphylococcal disease caused by community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA). The factors underlying the epidemic expansion of CA-MRSA lineages such as USA300, the predominant CA-MRSA clone in the United States, are largely unknown. Previously described virulence and antimicrobial resistance genes that promote the dissemination of CA-MRSA are carried by mobile genetic elements, including phages and plasmids. Here, we used high-resolution genomics and experimental infections to characterize the evolution of a USA300 variant plaguing a patient population at increased risk of infection to understand the mechanisms underlying the emergence of genetic elements that facilitate clonal spread of the pathogen. Genetic analyses provided conclusive evidence that fitness (manifest as emergence of a dominant clone) changed coincidently with the stepwise emergence of (i) a unique prophage and mutation of the regulator of the pyrimidine nucleotide biosynthetic operon that promoted abscess formation and colonization, respectively, thereby priming the clone for success; and (ii) a unique plasmid that conferred resistance to two topical microbiocides, mupirocin and chlorhexidine, frequently used for decolonization and infection prevention. The resistance plasmid evolved through successive incorporation of DNA elements from non-S. aureus spp. into an indigenous cryptic plasmid, suggesting a mechanism for interspecies genetic exchange that promotes antimicrobial resistance. Collectively, the data suggest that clonal spread in a vulnerable population resulted from extensive clinical intervention and intense selection pressure toward a pathogen lifestyle that involved the evolution of consequential mutations and mobile genetic elements.
Keywords: MRSA; antimicrobial resistance; evolution; virulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Ammerlaan HS, et al. Secular trends in nosocomial bloodstream infections: Antibiotic-resistant bacteria increase the total burden of infection. Clin Infect Dis. 2013;56:798–805. - PubMed
-
- Moran GJ, et al. EMERGEncy ID Net Study Group Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med. 2006;355:666–674. - PubMed
-
- Popovich KJ, Weinstein RA, Hota B. Are community-associated methicillin-resistant Staphylococcus aureus (MRSA) strains replacing traditional nosocomial MRSA strains? Clin Infect Dis. 2008;46:787–794. - PubMed
-
- Seybold U, et al. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care-associated blood stream infections. Clin Infect Dis. 2006;42:647–656. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI099394/AI/NIAID NIH HHS/United States
- S10 OD018522/OD/NIH HHS/United States
- R01 AI103268/AI/NIAID NIH HHS/United States
- HHSN272201400019C/AI/NIAID NIH HHS/United States
- T32 AI007180/AI/NIAID NIH HHS/United States
- S10 OD023423/OD/NIH HHS/United States
- KL2 TR001435/TR/NCATS NIH HHS/United States
- K08 AI101005/AI/NIAID NIH HHS/United States
- R01 AI119145/AI/NIAID NIH HHS/United States
- T32 AI007647/AI/NIAID NIH HHS/United States
- R01 AI137526/AI/NIAID NIH HHS/United States
- R01 AI105129/AI/NIAID NIH HHS/United States
- MR/R015600/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

