Crystal Structure of the Human Cannabinoid Receptor CB2
- PMID: 30639103
- PMCID: PMC6713262
- DOI: 10.1016/j.cell.2018.12.011
Crystal Structure of the Human Cannabinoid Receptor CB2
Abstract
The cannabinoid receptor CB2 is predominately expressed in the immune system, and selective modulation of CB2 without the psychoactivity of CB1 has therapeutic potential in inflammatory, fibrotic, and neurodegenerative diseases. Here, we report the crystal structure of human CB2 in complex with a rationally designed antagonist, AM10257, at 2.8 Å resolution. The CB2-AM10257 structure reveals a distinctly different binding pose compared with CB1. However, the extracellular portion of the antagonist-bound CB2 shares a high degree of conformational similarity with the agonist-bound CB1, which led to the discovery of AM10257's unexpected opposing functional profile of CB2 antagonism versus CB1 agonism. Further structural analysis using mutagenesis studies and molecular docking revealed the molecular basis of their function and selectivity for CB2 and CB1. Additional analyses of our designed antagonist and agonist pairs provide important insight into the activation mechanism of CB2. The present findings should facilitate rational drug design toward precise modulation of the endocannabinoid system.
Keywords: G-protein coupled receptor; cannabinoid receptor CB2; crystal structure; ligand design; subtype selectivity.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
All authors declare no competing interests.
Figures

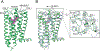
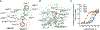
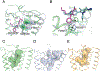


Comment in
-
Receptor Structures for a Caldron of Cannabinoids.Cell. 2019 Jan 24;176(3):409-411. doi: 10.1016/j.cell.2019.01.012. Cell. 2019. PMID: 30682366
Similar articles
-
Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-Gi Complex Structures.Cell. 2020 Feb 20;180(4):655-665.e18. doi: 10.1016/j.cell.2020.01.008. Epub 2020 Jan 30. Cell. 2020. PMID: 32004463 Free PMC article.
-
Prediction of the Binding Affinities and Selectivity for CB1 and CB2 Ligands Using Homology Modeling, Molecular Docking, Molecular Dynamics Simulations, and MM-PBSA Binding Free Energy Calculations.ACS Chem Neurosci. 2020 Apr 15;11(8):1139-1158. doi: 10.1021/acschemneuro.9b00696. Epub 2020 Apr 2. ACS Chem Neurosci. 2020. PMID: 32196303
-
Cryo-EM Structure of the Human Cannabinoid Receptor CB2-Gi Signaling Complex.Cell. 2020 Feb 20;180(4):645-654.e13. doi: 10.1016/j.cell.2020.01.007. Epub 2020 Jan 30. Cell. 2020. PMID: 32004460 Free PMC article.
-
Cannabinoid CB1 and CB2 receptor ligand specificity and the development of CB2-selective agonists.Curr Med Chem. 2008;15(14):1428-43. doi: 10.2174/092986708784567716. Curr Med Chem. 2008. PMID: 18537620 Review.
-
The pharmacology of cannabinoid receptors and their ligands: an overview.Int J Obes (Lond). 2006 Apr;30 Suppl 1:S13-8. doi: 10.1038/sj.ijo.0803272. Int J Obes (Lond). 2006. PMID: 16570099 Review.
Cited by
-
Understanding the Medical Chemistry of the Cannabis Plant is Critical to Guiding Real World Clinical Evidence.Molecules. 2020 Sep 4;25(18):4042. doi: 10.3390/molecules25184042. Molecules. 2020. PMID: 32899678 Free PMC article. Review.
-
Design, Synthesis, and Biological Activity of New CB2 Receptor Ligands: from Orthosteric and Allosteric Modulators to Dualsteric/Bitopic Ligands.J Med Chem. 2022 Jul 28;65(14):9918-9938. doi: 10.1021/acs.jmedchem.2c00582. Epub 2022 Jul 18. J Med Chem. 2022. PMID: 35849804 Free PMC article.
-
Discovery of 1,3-disubstituted pyrazole peripheral cannabinoid receptor partial agonists.Bioorg Med Chem Lett. 2023 Sep 1;93:129430. doi: 10.1016/j.bmcl.2023.129430. Epub 2023 Aug 4. Bioorg Med Chem Lett. 2023. PMID: 37543275 Free PMC article.
-
Placental Cannabinoid Receptor Expression in Preterm Birth.J Pregnancy. 2024 May 7;2024:6620156. doi: 10.1155/2024/6620156. eCollection 2024. J Pregnancy. 2024. PMID: 38745869 Free PMC article.
-
Positron Emission Tomography Imaging of the Endocannabinoid System: Opportunities and Challenges in Radiotracer Development.J Med Chem. 2021 Jan 14;64(1):123-149. doi: 10.1021/acs.jmedchem.0c01459. Epub 2020 Dec 30. J Med Chem. 2021. PMID: 33379862 Free PMC article. Review.
References
-
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, and Lindahl E (2015). GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

