Discovery of Novel Androgen Receptor Ligands by Structure-based Virtual Screening and Bioassays
- PMID: 30639122
- PMCID: PMC6411960
- DOI: 10.1016/j.gpb.2018.03.007
Discovery of Novel Androgen Receptor Ligands by Structure-based Virtual Screening and Bioassays
Abstract
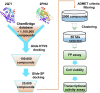
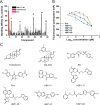
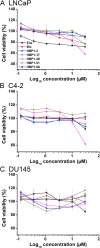
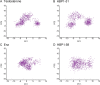
Androgen receptor (AR) is a ligand-activated transcription factor that plays a pivotal role in the development and progression of many severe diseases such as prostate cancer, muscle atrophy, and osteoporosis. Binding of ligands to AR triggers the conformational changes in AR that may affect the recruitment of coactivators and downstream response of AR signaling pathway. Therefore, AR ligands have great potential to treat these diseases. In this study, we searched for novel AR ligands by performing a docking-based virtual screening (VS) on the basis of the crystal structure of the AR ligand binding domain (LBD) in complex with its agonist. A total of 58 structurally diverse compounds were selected and subjected to LBD affinity assay, with five of them (HBP1-3, HBP1-17, HBP1-38, HBP1-51, and HBP1-58) exhibiting strong binding to AR-LBD. The IC50 values of HBP1-51 and HBP1-58 are 3.96 µM and 4.92 µM, respectively, which are even lower than that of enzalutamide (Enz, IC50 = 13.87 µM), a marketed second-generation AR antagonist. Further bioactivity assays suggest that HBP1-51 is an AR agonist, whereas HBP1-58 is an AR antagonist. In addition, molecular dynamics (MD) simulations and principal components analysis (PCA) were carried out to reveal the binding principle of the newly-identified AR ligands toward AR. Our modeling results indicate that the conformational changes of helix 12 induced by the bindings of antagonist and agonist are visibly different. In summary, the current study provides a highly efficient way to discover novel AR ligands, which could serve as the starting point for development of new therapeutics for AR-related diseases.
Keywords: AR agonist; AR antagonist; AR ligand; Androgen receptor; Virtual screening.
Copyright © 2019 The Authors. Production and hosting by Elsevier B.V. All rights reserved.
Figures





References
-
- Lu N.Z., Wardell S.E., Burnstein K.L., Defranco D., Fuller P.J., Giguere V. International Union of Pharmacology. LXV. The pharmacology and classification of the nuclear receptor superfamily: glucocorticoid, mineralocorticoid, progesterone, and androgen receptors. Pharmacol Rev. 2006;58:782–797. - PubMed
-
- Chen C.D., Welsbie D.S., Tran C., Baek S.H., Chen R., Vessella R. Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004;10:33–39. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

