The Surface Proteome of Adult Neural Stem Cells in Zebrafish Unveils Long-Range Cell-Cell Connections and Age-Related Changes in Responsiveness to IGF
- PMID: 30639211
- PMCID: PMC6373494
- DOI: 10.1016/j.stemcr.2018.12.005
The Surface Proteome of Adult Neural Stem Cells in Zebrafish Unveils Long-Range Cell-Cell Connections and Age-Related Changes in Responsiveness to IGF
Abstract
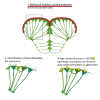
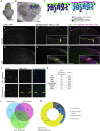
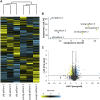
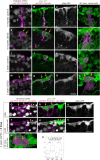
In adult stem cell populations, recruitment into division is parsimonious and most cells maintain a quiescent state. How individual cells decide to enter the cell cycle and how they coordinate their activity remains an essential problem to be resolved. It is thus important to develop methods to elucidate the mechanisms of cell communication and recruitment into the cell cycle. We made use of the advantageous architecture of the adult zebrafish telencephalon to isolate the surface proteins of an intact neural stem cell (NSC) population. We identified the proteome of NSCs in young and old brains. The data revealed a group of proteins involved in filopodia, which we validated by a morphological analysis of single cells, showing apically located cellular extensions. We further identified an age-related decrease in insulin-like growth factor (IGF) receptors. Expressing IGF2b induced divisions in young brains but resulted in incomplete divisions in old brains, stressing the role of cell-intrinsic processes in stem cell behavior.
Keywords: GFAP; aging; biotinylation; filopodia; lamellipodia; mass spectrometry; neurogenesis; pallium; quiescence; radial glia; telencephalon.
Copyright © 2018. Published by Elsevier Inc.
Figures



Similar articles
-
Reoccurring neural stem cell divisions in the adult zebrafish telencephalon are sufficient for the emergence of aggregated spatiotemporal patterns.PLoS Biol. 2020 Dec 8;18(12):e3000708. doi: 10.1371/journal.pbio.3000708. eCollection 2020 Dec. PLoS Biol. 2020. PMID: 33290409 Free PMC article.
-
Increased radial glia quiescence, decreased reactivation upon injury and unaltered neuroblast behavior underlie decreased neurogenesis in the aging zebrafish telencephalon.J Comp Neurol. 2013 Sep 1;521(13):3099-115. doi: 10.1002/cne.23347. J Comp Neurol. 2013. PMID: 23787922
-
Image analysis of neural stem cell division patterns in the zebrafish brain.Cytometry A. 2018 Mar;93(3):314-322. doi: 10.1002/cyto.a.23260. Epub 2017 Nov 10. Cytometry A. 2018. PMID: 29125897 Free PMC article.
-
Insulin and IGF receptor signalling in neural-stem-cell homeostasis.Nat Rev Endocrinol. 2015 Mar;11(3):161-70. doi: 10.1038/nrendo.2014.208. Epub 2014 Dec 2. Nat Rev Endocrinol. 2015. PMID: 25445849 Free PMC article. Review.
-
Neural Stem Cell Regulation in Zebrafish.Annu Rev Genet. 2024 Nov;58(1):249-272. doi: 10.1146/annurev-genet-111523-101949. Epub 2024 Nov 14. Annu Rev Genet. 2024. PMID: 39121542 Review.
Cited by
-
Reoccurring neural stem cell divisions in the adult zebrafish telencephalon are sufficient for the emergence of aggregated spatiotemporal patterns.PLoS Biol. 2020 Dec 8;18(12):e3000708. doi: 10.1371/journal.pbio.3000708. eCollection 2020 Dec. PLoS Biol. 2020. PMID: 33290409 Free PMC article.
-
Dynamic spatiotemporal coordination of neural stem cell fate decisions occurs through local feedback in the adult vertebrate brain.Cell Stem Cell. 2021 Aug 5;28(8):1457-1472.e12. doi: 10.1016/j.stem.2021.03.014. Epub 2021 Apr 5. Cell Stem Cell. 2021. PMID: 33823144 Free PMC article.
-
Glial Diversity and Evolution: Insights from Teleost Fish.Brain Sci. 2025 Jul 11;15(7):743. doi: 10.3390/brainsci15070743. Brain Sci. 2025. PMID: 40722334 Free PMC article. Review.
-
Protrusion-Mediated Signaling Regulates Patterning of the Developing Nervous System.Front Cell Dev Biol. 2020 Sep 29;8:579073. doi: 10.3389/fcell.2020.579073. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33134296 Free PMC article. Review.
-
Piezo1 regulates the regenerative capacity of skeletal muscles via orchestration of stem cell morphological states.Sci Adv. 2022 Mar 18;8(11):eabn0485. doi: 10.1126/sciadv.abn0485. Epub 2022 Mar 18. Sci Adv. 2022. PMID: 35302846 Free PMC article.
References
-
- Adolf B., Chapouton P., Lam C.S., Topp S., Tannhauser B., Strahle U., Gotz M., Bally-Cuif L. Conserved and acquired features of adult neurogenesis in the zebrafish telencephalon. Dev. Biol. 2006;295:278–293. - PubMed
-
- Barbosa J.S., Sanchez-Gonzalez R., Di Giaimo R., Baumgart E.V., Theis F.J., Gotz M., Ninkovic J. Neurodevelopment. Live imaging of adult neural stem cell behavior in the intact and injured zebrafish brain. Science. 2015;348:789–793. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

