Identification of candidate diagnostic and prognostic biomarkers for pancreatic carcinoma
- PMID: 30639415
- PMCID: PMC6412825
- DOI: 10.1016/j.ebiom.2019.01.003
Identification of candidate diagnostic and prognostic biomarkers for pancreatic carcinoma
Abstract
Background: Pancreatic carcinoma (PC) is one of the most aggressive cancers affecting human health. It is essential to identify candidate biomarkers for the diagnosis and prognosis of PC. The present study aimed to investigate the diagnosis and prognosis biomarkers of PC.
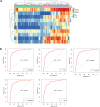
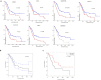
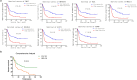
Methods: Differentially expressed genes (DEGs) were identified from the mRNA expression profiles of GSE62452, GSE28735 and GSE16515. Functional analysis and the protein-protein interaction network analysis was performed to explore the biological function of the identified DEGs. Diagnosis markers for PC were identified using ROC curve analysis. Prognosis markers were identified via survival analysis of TCGA data. The protein expression pattern of the identified genes was verified in clinical tissue samples. A retrospective clinical study was performed to evaluate the correlation between the expression of candidate proteins and survival time of patients. Moreover, comprehensive analysis of the combination of multiple genes/proteins for the prognosis prediction of PC was performed using both TCGA data and clinical data. In vitro studies were undertaken to elaborate the potential roles of these biomarkers in clonability and invasion of PC cells.
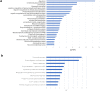
Findings: In total, 389 DEGs were identified. These genes were mainly associated with pancreatic secretion, protein digestion and absorption, cytochrome P450 drug metabolism, and energy metabolism pathway. The top 10 genes were filtered out following Fisher's exact test. ROC curve analysis demonstrated that TMPRSS4, SERPINB5, SLC6A14, SCEL, and TNS4 could be used as biomarkers for the diagnosis of PC. Survival analysis of TCGA data and clinical data suggested that TMC7, TMPRSS4, SCEL, SLC2A1, CENPF, SERPINB5 and SLC6A14 can be potential biomarkers for the prognosis of PC. Comprehensive analysis show that a combination of identified genes/proteins can predict the prognosis of PC. Mechanistically, the identified genes attributes to clonability and invasiveness of PC cells.
Interpretation: We synthesized several sets of public data and preliminarily clarified pathways and functions of PC. Candidate molecular markers were identified for diagnosis and prognosis prediction of PC including a novel gene, TMC7. Moreover, we found that the combination of TMC7, TMPRSS4, SCEL, SLC2A1, CENPF, SERPINB5 and SLC6A14 can serve as a promising indicator of the prognosis of PC patients. The candidate proteins may attribute to clonability and invasiveness of PC cells. This research provides a novel insight into molecular mechanisms as well as diagnostic and prognostic markers of PC. FUND: National Natural Science Foundation of China [No. 81602646 &81802339], Natural Science Foundation of Guangdong Province [No. 2016A030310254] and China Postdoctoral Science Foundation [No. 2016M600648].
Keywords: Biomarker; Diagnosis; Function; Pancreatic carcinoma; Prognosis.
Copyright © 2019. Published by Elsevier B.V.
Figures



References
-
- Abel E.V., Simeone D.M. Biology and clinical applications of pancreatic cancer stem cells. Gastroenterology. 2013 Jun;144(6):1241–1248. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

