Transfer Learning with Deep Convolutional Neural Networks for Classifying Cellular Morphological Changes
- PMID: 30641024
- PMCID: PMC6484664
- DOI: 10.1177/2472555218818756
Transfer Learning with Deep Convolutional Neural Networks for Classifying Cellular Morphological Changes
Abstract
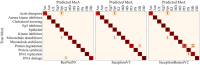
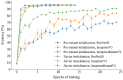
The quantification and identification of cellular phenotypes from high-content microscopy images has proven to be very useful for understanding biological activity in response to different drug treatments. The traditional approach has been to use classical image analysis to quantify changes in cell morphology, which requires several nontrivial and independent analysis steps. Recently, convolutional neural networks have emerged as a compelling alternative, offering good predictive performance and the possibility to replace traditional workflows with a single network architecture. In this study, we applied the pretrained deep convolutional neural networks ResNet50, InceptionV3, and InceptionResnetV2 to predict cell mechanisms of action in response to chemical perturbations for two cell profiling datasets from the Broad Bioimage Benchmark Collection. These networks were pretrained on ImageNet, enabling much quicker model training. We obtain higher predictive accuracy than previously reported, between 95% and 97%. The ability to quickly and accurately distinguish between different cell morphologies from a scarce amount of labeled data illustrates the combined benefit of transfer learning and deep convolutional neural networks for interrogating cell-based images.
Keywords: cell phenotypes; deep learning; high-content imaging; machine learning; transfer learning.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

