Metabolic reprogramming of oncogene-addicted cancer cells to OXPHOS as a mechanism of drug resistance
- PMID: 30642723
- PMCID: PMC6859574
- DOI: 10.1016/j.redox.2018.101076
Metabolic reprogramming of oncogene-addicted cancer cells to OXPHOS as a mechanism of drug resistance
Abstract
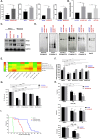
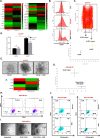
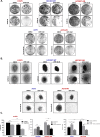
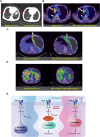
The ability to selectively eradicate oncogene-addicted tumors while reducing systemic toxicity has endeared targeted therapies as a treatment strategy. Nevertheless, development of acquired resistance limits the benefits and durability of such a regime. Here we report evidence of enhanced reliance on mitochondrial oxidative phosphorylation (OXPHOS) in oncogene-addicted cancers manifesting acquired resistance to targeted therapies. To that effect, we describe a novel OXPHOS targeting activity of the small molecule compound, OPB-51602 (OPB). Of note, a priori treatment with OPB restored sensitivity to targeted therapies. Furthermore, cancer cells exhibiting stemness markers also showed selective reliance on OXPHOS and enhanced sensitivity to OPB. Importantly, in a subset of patients who developed secondary resistance to EGFR tyrosine kinase inhibitor (TKI), OPB treatment resulted in decrease in metabolic activity and reduction in tumor size. Collectively, we show here a switch to mitochondrial OXPHOS as a key driver of targeted drug resistance in oncogene-addicted cancers. This metabolic vulnerability is exploited by a novel OXPHOS inhibitor, which also shows promise in the clinical setting.
Keywords: Metabolic reprogramming; OXPHOS; Oncogene-addiction; STAT3.
Copyright © 2018 The Authors. Published by Elsevier B.V. All rights reserved.
Figures


References
-
- Ashton T.M., McKenna W.G., Kunz-Schughart L.A., Higgins G.S. Oxidative phosphorylation as an emerging target in cancer therapy. Clin. Cancer Res. 2018 - PubMed
-
- Jose C., Bellance N., Rossignol R. Choosing between glycolysis and oxidative phosphorylation: a tumor's dilemma? Biochim. Biophys. Acta. 2011;1807:552–561. - PubMed
-
- Solaini G., Sgarbi G., Baracca A. Oxidative phosphorylation in cancer cells. Biochim. Biophys. Acta. 2011;1807:534–542. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

