Flagellar Stators Stimulate c-di-GMP Production by Pseudomonas aeruginosa
- PMID: 30642992
- PMCID: PMC6707927
- DOI: 10.1128/JB.00741-18
Flagellar Stators Stimulate c-di-GMP Production by Pseudomonas aeruginosa
Abstract
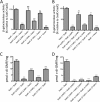
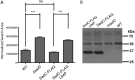
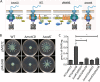
Flagellar motility is critical for surface attachment and biofilm formation in many bacteria. A key regulator of flagellar motility in Pseudomonas aeruginosa and other microbes is cyclic diguanylate (c-di-GMP). High levels of this second messenger repress motility and stimulate biofilm formation. c-di-GMP levels regulate motility in P. aeruginosa in part by influencing the localization of its two flagellar stator sets, MotAB and MotCD. Here, we show that while c-di-GMP can influence stator localization, stators can in turn impact c-di-GMP levels. We demonstrate that the swarming motility-driving stator MotC physically interacts with the transmembrane region of the diguanylate cyclase SadC. Furthermore, we demonstrate that this interaction is capable of stimulating SadC activity. We propose a model by which the MotCD stator set interacts with SadC to stimulate c-di-GMP production under conditions not permissive to motility. This regulation implies a positive-feedback loop in which c-di-GMP signaling events cause MotCD stators to disengage from the motor; then disengaged stators stimulate c-di-GMP production to reinforce a biofilm mode of growth. Our studies help to define the bidirectional interactions between c-di-GMP and the flagellar machinery.IMPORTANCE The ability of bacterial cells to control motility during early steps in biofilm formation is critical for the transition to a nonmotile, biofilm lifestyle. Recent studies have clearly demonstrated the ability of c-di-GMP to control motility via a number of mechanisms, including through controlling transcription of motility-related genes and modulating motor function. Here, we provide evidence that motor components can in turn impact c-di-GMP levels. We propose that communication between motor components and the c-di-GMP synthesis machinery allows the cell to have a robust and sensitive switching mechanism to control motility during early events in biofilm formation.
Keywords: Pseudomonas aeruginosa; biofilm; c-di-GMP; flagella; motility; stator.
Copyright © 2019 American Society for Microbiology.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

