Strigolactone perception and deactivation by a hydrolase receptor DWARF14
- PMID: 30643123
- PMCID: PMC6331613
- DOI: 10.1038/s41467-018-08124-7
Strigolactone perception and deactivation by a hydrolase receptor DWARF14
Abstract
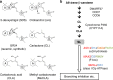
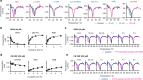
The perception mechanism for the strigolactone (SL) class of plant hormones has been a subject of debate because their receptor, DWARF14 (D14), is an α/β-hydrolase that can cleave SLs. Here we show via time-course analyses of SL binding and hydrolysis by Arabidopsis thaliana D14, that the level of uncleaved SL strongly correlates with the induction of the active signaling state. In addition, we show that an AtD14D218A catalytic mutant that lacks enzymatic activity is still able to complement the atd14 mutant phenotype in an SL-dependent manner. We conclude that the intact SL molecules trigger the D14 active signaling state, and we also describe that D14 deactivates bioactive SLs by the hydrolytic degradation after signal transmission. Together, these results reveal that D14 is a dual-functional receptor, responsible for both the perception and deactivation of bioactive SLs.
Conflict of interest statement
The authors declare no competing interests.
Figures



Comment in
-
Binding or Hydrolysis? How Does the Strigolactone Receptor Work?Trends Plant Sci. 2019 Jul;24(7):571-574. doi: 10.1016/j.tplants.2019.05.001. Epub 2019 May 28. Trends Plant Sci. 2019. PMID: 31151745
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

