Altered spinogenesis in iPSC-derived cortical neurons from patients with autism carrying de novo SHANK3 mutations
- PMID: 30643170
- PMCID: PMC6331634
- DOI: 10.1038/s41598-018-36993-x
Altered spinogenesis in iPSC-derived cortical neurons from patients with autism carrying de novo SHANK3 mutations
Abstract
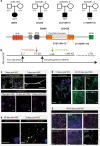
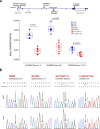
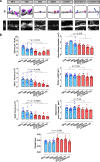
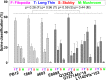
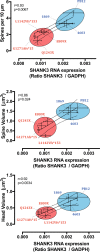
The synaptic protein SHANK3 encodes a multidomain scaffold protein expressed at the postsynaptic density of neuronal excitatory synapses. We previously identified de novo SHANK3 mutations in patients with autism spectrum disorders (ASD) and showed that SHANK3 represents one of the major genes for ASD. Here, we analyzed the pyramidal cortical neurons derived from induced pluripotent stem cells from four patients with ASD carrying SHANK3 de novo truncating mutations. At 40-45 days after the differentiation of neural stem cells, dendritic spines from pyramidal neurons presented variable morphologies: filopodia, thin, stubby and muschroom, as measured in 3D using GFP labeling and immunofluorescence. As compared to three controls, we observed a significant decrease in SHANK3 mRNA levels (less than 50% of controls) in correlation with a significant reduction in dendritic spine densities and whole spine and spine head volumes. These results, obtained through the analysis of de novo SHANK3 mutations in the patients' genomic background, provide further support for the presence of synaptic abnormalities in a subset of patients with ASD.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

