Astrocytoma progression scoring system based on the WHO 2016 criteria
- PMID: 30643174
- PMCID: PMC6331604
- DOI: 10.1038/s41598-018-36471-4
Astrocytoma progression scoring system based on the WHO 2016 criteria
Abstract
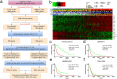
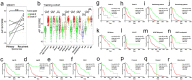
Diffuse astrocytoma (including glioblastoma) is morbid with a worse prognosis than other types of glioma. Therefore, we sought to build a progression-associated score to improve malignancy and prognostic predictions for astrocytoma. The astrocytoma progression (AP) score was constructed through bioinformatics analyses of the training cohort (TCGA RNA-seq) and included 18 genes representing distinct aspects of regulation during astrocytoma progression. This classifier could successfully discriminate patients with distinct prognoses in the training and validation (REMBRANDT, GSE16011 and TCGA-GBM Microarray) cohorts (P < 0.05 in all cohorts) and in different clinicopathological subgroups. Distinct patterns of somatic mutations and copy number variation were also observed. The bioinformatics analyses suggested that genes associated with a higher AP score were significantly involved in cancer progression-related biological processes, such as the cell cycle and immune/inflammatory responses, whereas genes associated with a lower AP score were associated with relatively normal nervous system biological processes. The analyses indicated that the AP score was a robust predictor of patient survival, and its ability to predict astrocytoma malignancy was well elucidated. Therefore, this bioinformatics-based scoring system suggested that astrocytoma progression could distinguish patients with different underlying biological processes and clinical outcomes, facilitate more precise tumour grading and possibly shed light on future classification strategies and therapeutics for astrocytoma patients.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Xing Z, You RX, Li J, Liu Y, Cao DR. Differentiation of primary central nervous system lymphomas from high-grade gliomas by rCBV and percentage of signal intensity recovery derived from dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. Clin Neuroradiol. 2014;24:329–336. doi: 10.1007/s00062-013-0255-5. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

