Cancer therapeutic targeting using mutant-p53-specific siRNAs
- PMID: 30643191
- PMCID: PMC6756012
- DOI: 10.1038/s41388-018-0652-y
Cancer therapeutic targeting using mutant-p53-specific siRNAs
Abstract
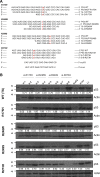
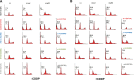
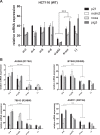
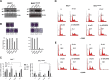
Mutations in Tp53 compromise therapeutic response, due either to the dominant-negative effect over the functional wild-type allele; or as a result of the survival advantage conferred by mutant p53 to which cancer cells become addicted. Thus, targeting mutant p53 represents an effective therapeutic strategy to treat over half of all cancers. We have therefore generated a series of small-interfering-RNAs, capable of targeting four p53 hot-spot mutants which represent ~20% of all p53 mutations. These mutant-p53-specific siRNAs (MupSi) are highly specific in silencing the expression of the intended mutants without affecting wild-type p53. Functionally, these MupSis induce cell death by abrogating both the addiction to mutant p53 and the dominant-negative effect; and retard tumor growth in xenografts when administered in a therapeutic setting. These data together demonstrate the possibility of targeting mutant p53 specifically to improve clinical outcome.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
Allele-specific silencing of mutant p53 attenuates dominant-negative and gain-of-function activities.Oncotarget. 2016 Feb 2;7(5):5401-15. doi: 10.18632/oncotarget.6634. Oncotarget. 2016. PMID: 26700961 Free PMC article.
-
Dominant-negative p53 mutant R248Q increases the motile and invasive activities of oral squamous cell carcinoma cells.Biomed Res. 2019;40(1):37-49. doi: 10.2220/biomedres.40.37. Biomed Res. 2019. PMID: 30787262
-
Specific TP53 Mutants Overrepresented in Ovarian Cancer Impact CNV, TP53 Activity, Responses to Nutlin-3a, and Cell Survival.Neoplasia. 2015 Oct;17(10):789-803. doi: 10.1016/j.neo.2015.10.003. Neoplasia. 2015. PMID: 26585234 Free PMC article.
-
Targeting mutant p53 for cancer therapy: direct and indirect strategies.J Hematol Oncol. 2021 Sep 28;14(1):157. doi: 10.1186/s13045-021-01169-0. J Hematol Oncol. 2021. PMID: 34583722 Free PMC article. Review.
-
Mutant p53: it's not all one and the same.Cell Death Differ. 2022 May;29(5):983-987. doi: 10.1038/s41418-022-00989-y. Epub 2022 Mar 31. Cell Death Differ. 2022. PMID: 35361963 Free PMC article. Review.
Cited by
-
Targeting Mutant p53 for Cancer Treatment: Moving Closer to Clinical Use?Cancers (Basel). 2022 Sep 16;14(18):4499. doi: 10.3390/cancers14184499. Cancers (Basel). 2022. PMID: 36139658 Free PMC article. Review.
-
Role of p53 in breast cancer progression: An insight into p53 targeted therapy.Theranostics. 2023 Feb 27;13(4):1421-1442. doi: 10.7150/thno.81847. eCollection 2023. Theranostics. 2023. PMID: 36923534 Free PMC article. Review.
-
Recent advances in targeting the "undruggable" proteins: from drug discovery to clinical trials.Signal Transduct Target Ther. 2023 Sep 6;8(1):335. doi: 10.1038/s41392-023-01589-z. Signal Transduct Target Ther. 2023. PMID: 37669923 Free PMC article. Review.
-
The Effects of Single Nucleotide Polymorphisms in Cancer RNAi Therapies.Cancers (Basel). 2020 Oct 25;12(11):3119. doi: 10.3390/cancers12113119. Cancers (Basel). 2020. PMID: 33113880 Free PMC article. Review.
-
Mutant p53 Gain of Function: Why Many See It, Why Some Do Not.Cancer Discov. 2025 Jun 3;15(6):1099-1104. doi: 10.1158/2159-8290.CD-24-1638. Cancer Discov. 2025. PMID: 40287981
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

