PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia
- PMID: 30643249
- PMCID: PMC6525306
- DOI: 10.1038/s41588-018-0315-5
PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia
Abstract
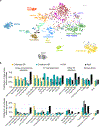
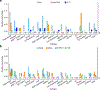
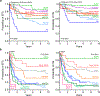
Recent genomic studies have identified chromosomal rearrangements defining new subtypes of B-progenitor acute lymphoblastic leukemia (B-ALL), however many cases lack a known initiating genetic alteration. Using integrated genomic analysis of 1,988 childhood and adult cases, we describe a revised taxonomy of B-ALL incorporating 23 subtypes defined by chromosomal rearrangements, sequence mutations or heterogeneous genomic alterations, many of which show marked variation in prevalence according to age. Two subtypes have frequent alterations of the B lymphoid transcription-factor gene PAX5. One, PAX5alt (7.4%), has diverse PAX5 alterations (rearrangements, intragenic amplifications or mutations); a second subtype is defined by PAX5 p.Pro80Arg and biallelic PAX5 alterations. We show that p.Pro80Arg impairs B lymphoid development and promotes the development of B-ALL with biallelic Pax5 alteration in vivo. These results demonstrate the utility of transcriptome sequencing to classify B-ALL and reinforce the central role of PAX5 as a checkpoint in B lymphoid maturation and leukemogenesis.
Conflict of interest statement
COMPETING INTERESTS
The authors declare no competing financial interests.
Figures




References
-
- Hunger SP & Mullighan CG Acute Lymphoblastic Leukemia in Children. N Engl J Med 373, 1541–52 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA016672/CA/NCI NIH HHS/United States
- R35 CA197695/CA/NCI NIH HHS/United States
- UG1 CA189859/CA/NCI NIH HHS/United States
- U24 CA196172/CA/NCI NIH HHS/United States
- P30 CA021765/CA/NCI NIH HHS/United States
- U10 CA180790/CA/NCI NIH HHS/United States
- P30 CA016058/CA/NCI NIH HHS/United States
- U10 CA180899/CA/NCI NIH HHS/United States
- UG1 CA233332/CA/NCI NIH HHS/United States
- U10 CA180886/CA/NCI NIH HHS/United States
- P50 GM115279/GM/NIGMS NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- U10 CA180791/CA/NCI NIH HHS/United States
- RC1 CA145707/CA/NCI NIH HHS/United States
- U10 CA180820/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

