Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use
- PMID: 30643251
- PMCID: PMC6358542
- DOI: 10.1038/s41588-018-0307-5
Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use
Abstract
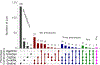
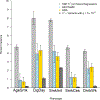
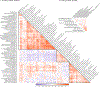
Tobacco and alcohol use are leading causes of mortality that influence risk for many complex diseases and disorders1. They are heritable2,3 and etiologically related4,5 behaviors that have been resistant to gene discovery efforts6-11. In sample sizes up to 1.2 million individuals, we discovered 566 genetic variants in 406 loci associated with multiple stages of tobacco use (initiation, cessation, and heaviness) as well as alcohol use, with 150 loci evidencing pleiotropic association. Smoking phenotypes were positively genetically correlated with many health conditions, whereas alcohol use was negatively correlated with these conditions, such that increased genetic risk for alcohol use is associated with lower disease risk. We report evidence for the involvement of many systems in tobacco and alcohol use, including genes involved in nicotinic, dopaminergic, and glutamatergic neurotransmission. The results provide a solid starting point to evaluate the effects of these loci in model organisms and more precise substance use measures.
Conflict of interest statement
Figures

References
-
- Ezzati M et al. Selected major risk factors and global and regional burden of disease. Lancet 360, 1347–1360 (2002). - PubMed
-
- Polderman TJ et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet (2015). - PubMed
-
- Kendler KS, Prescott CA, Myers J & Neale MC The structure of genetic and environmental risk factors for common psychiatric and substance use disorders in men and women. Archives of General Psychiatry 60, 929–937 (2003). - PubMed
METHODS ONLY REFERENCES
-
- Price AL et al. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics 38, 904–909 (2006). - PubMed
Publication types
MeSH terms
Grants and funding
- R01 DK093757/DK/NIDDK NIH HHS/United States
- R01 HL089897/HL/NHLBI NIH HHS/United States
- P01 HD031921/HD/NICHD NIH HHS/United States
- K01 HL125858/HL/NHLBI NIH HHS/United States
- U01 AG009740/AG/NIA NIH HHS/United States
- R03 OD032630/OD/NIH HHS/United States
- R01 GM126479/GM/NIGMS NIH HHS/United States
- R03 HD097630/HD/NICHD NIH HHS/United States
- R01 DA042755/DA/NIDA NIH HHS/United States
- G0902144/MRC_/Medical Research Council/United Kingdom
- P2C HD066613/HD/NICHD NIH HHS/United States
- T32 DK091317/DK/NIDDK NIH HHS/United States
- K01 MH109765/MH/NIMH NIH HHS/United States
- R01 MH123619/MH/NIMH NIH HHS/United States
- R03 DC013373/DC/NIDCD NIH HHS/United States
- R21 DA040177/DA/NIDA NIH HHS/United States
- U01 HL089897/HL/NHLBI NIH HHS/United States
- MC_UU_00011/7/MRC_/Medical Research Council/United Kingdom
- R01 DK072193/DK/NIDDK NIH HHS/United States
- MR/K023195/1/MRC_/Medical Research Council/United Kingdom
- T32 HG000040/HG/NHGRI NIH HHS/United States
- R01 HL119443/HL/NHLBI NIH HHS/United States
- R01 DK062370/DK/NIDDK NIH HHS/United States
- U01 HL089856/HL/NHLBI NIH HHS/United States
- R01 DA034076/DA/NIDA NIH HHS/United States
- R01 HG008983/HG/NHGRI NIH HHS/United States
- R01 DA036583/DA/NIDA NIH HHS/United States
- MC_UU_12013/1/MRC_/Medical Research Council/United Kingdom
- U01 DK062370/DK/NIDDK NIH HHS/United States
- R21 AA021223/AA/NIAAA NIH HHS/United States
- R01 DA037904/DA/NIDA NIH HHS/United States
- U01 DA046413/DA/NIDA NIH HHS/United States
- R01 AA023974/AA/NIAAA NIH HHS/United States
- P30 DK020572/DK/NIDDK NIH HHS/United States
- S10 OD020069/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

