Regulatory T cells mediate specific suppression by depleting peptide-MHC class II from dendritic cells
- PMID: 30643268
- PMCID: PMC6402611
- DOI: 10.1038/s41590-018-0280-2
Regulatory T cells mediate specific suppression by depleting peptide-MHC class II from dendritic cells
Abstract
Regulatory T cells (Treg cells) can activate multiple suppressive mechanisms in vitro after activation via the T cell antigen receptor, resulting in antigen-independent suppression. However, it remains unclear whether similar pathways operate in vivo. Here we found that antigen-specific Treg cells activated by dendritic cells (DCs) pulsed with two antigens suppressed conventional naive T cells (Tnaive cells) specific for both cognate antigens and non-cognate antigens in vitro but suppressed only Tnaive cells specific for cognate antigen in vivo. Antigen-specific Treg cells formed strong interactions with DCs, resulting in selective inhibition of the binding of Tnaive cells to cognate antigen yet allowing bystander Tnaive cell access. Strong binding resulted in the removal of the complex of cognate peptide and major histocompatibility complex class II (pMHCII) from the DC surface, reducing the capacity of DCs to present antigen. The enhanced binding of Treg cells to DCs, coupled with their capacity to deplete pMHCII, represents a novel pathway for Treg cell-mediated suppression and may be a mechanism by which Treg cells maintain immune homeostasis.
Conflict of interest statement
Competing interests
The authors declare no competing interests
Figures


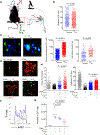
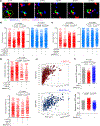
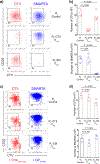
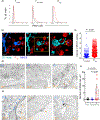


References
-
- Borsellino G et al. Expression of ectonucleotidase CD39 by Foxp3+ Treg cells: hydrolysis of extracellular ATP and immune suppression. Blood 110, 1225–1232 (2007). - PubMed
-
- Collison LW et al. The inhibitory cytokine IL-35 contributes to regulatory T-cell function. Nature 450, 566–569 (2007). - PubMed
-
- Shevach EM Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity 30, 636–645 (2009). - PubMed
REFERENCES FOR METHODS
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

