Discovery of a ZIP7 inhibitor from a Notch pathway screen
- PMID: 30643281
- PMCID: PMC7251565
- DOI: 10.1038/s41589-018-0200-7
Discovery of a ZIP7 inhibitor from a Notch pathway screen
Abstract
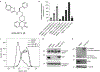
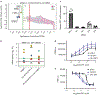
The identification of activating mutations in NOTCH1 in 50% of T cell acute lymphoblastic leukemia has generated interest in elucidating how these mutations contribute to oncogenic transformation and in targeting the pathway. A phenotypic screen identified compounds that interfere with trafficking of Notch and induce apoptosis via an endoplasmic reticulum (ER) stress mechanism. Target identification approaches revealed a role for SLC39A7 (ZIP7), a zinc transport family member, in governing Notch trafficking and signaling. Generation and sequencing of a compound-resistant cell line identified a V430E mutation in ZIP7 that confers transferable resistance to the compound NVS-ZP7-4. NVS-ZP7-4 altered zinc in the ER, and an analog of the compound photoaffinity labeled ZIP7 in cells, suggesting a direct interaction between the compound and ZIP7. NVS-ZP7-4 is the first reported chemical tool to probe the impact of modulating ER zinc levels and investigate ZIP7 as a novel druggable node in the Notch pathway.
Conflict of interest statement
Competing interests
E.N., S.G., L.L., S.B., S.M. Brittain, P.B.-E., J.J.L., J.R.T., M.S., Y.Y., N.G., G.R., S.S., M.B., A.L., F.S., A.C., K.X.X., S.H., J.R.-H., W.A.W., K.T., D.H., R.I.M., N.G., A.D., H.G., J.S., J.D., S.M.C., G.B., E.L.G., Z.B.K., C.A., J.A.P., O.W., J.A.T., J.L.J., R.K.J., S.M. Bushell, and C.J.F. are (or were at the time the research was conducted) employees of Novartis.
Figures




References
-
- Imming P, Sinning C & Meyer A Drugs, their targets and the nature and number of drug targets. Nat. Rev. Drug Discov 5, 821–834 (2006). - PubMed
-
- Overington JP, Al-Lazikani B & Hopkins AL How many drug targets are there? Nat. Rev. Drug Discov 5, 993–996 (2006). - PubMed
-
- Weng AP et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306, 269–271 (2004). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

