Translation control of the immune checkpoint in cancer and its therapeutic targeting
- PMID: 30643286
- PMCID: PMC6613562
- DOI: 10.1038/s41591-018-0321-2
Translation control of the immune checkpoint in cancer and its therapeutic targeting
Abstract
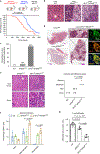
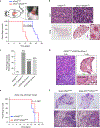
Cancer cells develop mechanisms to escape immunosurveillance, among which modulating the expression of immune suppressive messenger RNAs is most well-documented. However, how this is molecularly achieved remains largely unresolved. Here, we develop an in vivo mouse model of liver cancer to study oncogene cooperation in immunosurveillance. We show that MYC overexpression (MYCTg) synergizes with KRASG12D to induce an aggressive liver tumor leading to metastasis formation and reduced mouse survival compared with KRASG12D alone. Genome-wide ribosomal footprinting of MYCTg;KRASG12 tumors compared with KRASG12D revealed potential alterations in translation of mRNAs, including programmed-death-ligand 1 (PD-L1). Further analysis revealed that PD-L1 translation is repressed in KRASG12D tumors by functional, non-canonical upstream open reading frames in its 5' untranslated region, which is bypassed in MYCTg;KRASG12D tumors to evade immune attack. We show that this mechanism of PD-L1 translational upregulation was effectively targeted by a potent, clinical compound that inhibits eIF4E phosphorylation, eFT508, which reverses the aggressive and metastatic characteristics of MYCTg;KRASG12D tumors. Together, these studies reveal how immune-checkpoint proteins are manipulated by distinct oncogenes at the level of mRNA translation, which can be exploited for new immunotherapies.
Figures



References
-
- Chen DS & Mellman I Oncology meets immunology: the cancer-immunity cycle. Immunity 39, 1–10 (2013). - PubMed
-
- Stewart BW & Wild C World Cancer Report 2014 (International Agency for Research on Cancer WHO Press, 2014).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

