Healthy infants harbor intestinal bacteria that protect against food allergy
- PMID: 30643289
- PMCID: PMC6408964
- DOI: 10.1038/s41591-018-0324-z
Healthy infants harbor intestinal bacteria that protect against food allergy
Abstract
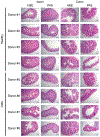
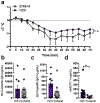
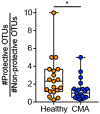
There has been a striking generational increase in life-threatening food allergies in Westernized societies1,2. One hypothesis to explain this rising prevalence is that twenty-first century lifestyle practices, including misuse of antibiotics, dietary changes, and higher rates of Caesarean birth and formula feeding have altered intestinal bacterial communities; early-life alterations may be particularly detrimental3,4. To better understand how commensal bacteria regulate food allergy in humans, we colonized germ-free mice with feces from healthy or cow's milk allergic (CMA) infants5. We found that germ-free mice colonized with bacteria from healthy, but not CMA, infants were protected against anaphylactic responses to a cow's milk allergen. Differences in bacterial composition separated the healthy and CMA populations in both the human donors and the colonized mice. Healthy and CMA colonized mice also exhibited unique transcriptome signatures in the ileal epithelium. Correlation of ileal bacteria with genes upregulated in the ileum of healthy or CMA colonized mice identified a clostridial species, Anaerostipes caccae, that protected against an allergic response to food. Our findings demonstrate that intestinal bacteria are critical for regulating allergic responses to dietary antigens and suggest that interventions that modulate bacterial communities may be therapeutically relevant for food allergy.
Figures

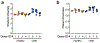
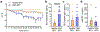


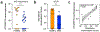


Comment in
-
Food allergy: could the gut microbiota hold the key?Nat Rev Gastroenterol Hepatol. 2019 Apr;16(4):201-202. doi: 10.1038/s41575-019-0123-0. Nat Rev Gastroenterol Hepatol. 2019. PMID: 30824883 No abstract available.
References
-
- Iweala OI & Burks AW Food Allergy: Our Evolving Understanding of Its Pathogenesis, Prevention, and Treatment. Curr Allergy Asthma Rep 16, 37 (2016). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

