ALS-implicated protein TDP-43 sustains levels of STMN2, a mediator of motor neuron growth and repair
- PMID: 30643292
- PMCID: PMC7153761
- DOI: 10.1038/s41593-018-0300-4
ALS-implicated protein TDP-43 sustains levels of STMN2, a mediator of motor neuron growth and repair
Abstract
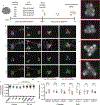
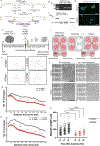
The findings that amyotrophic lateral sclerosis (ALS) patients almost universally display pathological mislocalization of the RNA-binding protein TDP-43 and that mutations in its gene cause familial ALS have nominated altered RNA metabolism as a disease mechanism. However, the RNAs regulated by TDP-43 in motor neurons and their connection to neuropathy remain to be identified. Here we report transcripts whose abundances in human motor neurons are sensitive to TDP-43 depletion. Notably, expression of STMN2, which encodes a microtubule regulator, declined after TDP-43 knockdown and TDP-43 mislocalization as well as in patient-specific motor neurons and postmortem patient spinal cord. STMN2 loss upon reduced TDP-43 function was due to altered splicing, which is functionally important, as we show STMN2 is necessary for normal axonal outgrowth and regeneration. Notably, post-translational stabilization of STMN2 rescued neurite outgrowth and axon regeneration deficits induced by TDP-43 depletion. We propose that restoring STMN2 expression warrants examination as a therapeutic strategy for ALS.
Conflict of interest statement
Competing Financial Interests
The authors have no competing financial interests.
Figures






References
-
- Miller RG, Mitchell JD & Moore DH Riluzole for amyotrophic lateral sclerosis (ALS)/motor neuron disease (MND). Cochrane Database Syst Rev 3, CD001447 (2012).
-
- Neumann M et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 (2006). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

