6'-Fluoro[4.3.0]bicyclo nucleic acid: synthesis, biophysical properties and molecular dynamics simulations
- PMID: 30643586
- PMCID: PMC6317435
- DOI: 10.3762/bjoc.14.288
6'-Fluoro[4.3.0]bicyclo nucleic acid: synthesis, biophysical properties and molecular dynamics simulations
Abstract
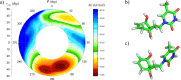
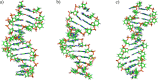
Here we report on the synthesis, biophysical properties and molecular modeling of oligonucleotides containing unsaturated 6'-fluoro[4.3.0]bicyclo nucleotides (6'F-bc4,3-DNA). Two 6'F-bc4,3 phosphoramidite building blocks (T and C) were synthesized starting from a previously described [3.3.0]bicyclic sugar. The conversion of this sugar to a gem-difluorinated tricyclic intermediate via difluorocarbene addition followed either by a NIS-mediated or Vorbrüggen nucleosidation yielded in both cases the β-tricyclic nucleoside as major anomer. Subsequent desilylation and cyclopropane ring opening of these tricyclic intermediates afforded the unsaturated 6'F-bc4,3 nucleosides. The successful incorporation of the corresponding phosphoramidite building blocks into oligonucleotides was achieved with tert-butyl hydroperoxide as oxidation agent. Thermal melting experiments of the modified duplexes disclosed a destabilizing effect versus DNA and RNA complements, but with a lesser degree of destabilization versus complementary DNA (ΔT m/mod = -1.5 to -3.7 °C). Molecular dynamics simulation on the nucleoside and oligonucleotide level revealed the preference of the C1'-exo/C2'-endo alignment of the furanose ring. Moreover, the simulation of duplexes with complementary RNA disclosed a DNA/RNA-type duplex structure suggesting that this modification might be a substrate for RNase H.
Keywords: DNA/RNA affinity; fluorinated cyclopropanes; fluorinated nucleic acids; molecular dynamics simulations; sugar modified nucleosides.
Figures






References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
