Probe dependence of allosteric enhancers on the binding affinity of adenosine A1 -receptor agonists at rat and human A1 -receptors measured using NanoBRET
- PMID: 30644086
- PMCID: PMC6433648
- DOI: 10.1111/bph.14575
Probe dependence of allosteric enhancers on the binding affinity of adenosine A1 -receptor agonists at rat and human A1 -receptors measured using NanoBRET
Abstract
Background and purpose: Adenosine is a local mediator that regulates a number of physiological and pathological processes via activation of adenosine A1 -receptors. The activity of adenosine can be regulated at the level of its target receptor via drugs that bind to an allosteric site on the A1 -receptor. Here, we have investigated the species and probe dependence of two allosteric modulators on the binding characteristics of fluorescent and nonfluorescent A1 -receptor agonists.
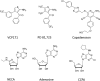
Experimental approach: A Nano-luciferase (Nluc) BRET (NanoBRET) methodology was used. This used N-terminal Nluc-tagged A1 -receptors expressed in HEK293T cells in conjunction with both fluorescent A1 -receptor agonists (adenosine and NECA analogues) and a fluorescent antagonist CA200645.
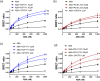
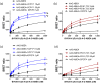
Key results: PD 81,723 and VCP171 elicited positive allosteric effects on the binding affinity of orthosteric agonists at both the rat and human A1 -receptors that showed clear probe dependence. Thus, the allosteric effect on the highly selective partial agonist capadenoson was much less marked than for the full agonists NECA, adenosine, and CCPA in both species. VCP171 and, to a lesser extent, PD 81,723, also increased the specific binding of three fluorescent A1 -receptor agonists in a species-dependent manner that involved increases in Bmax and pKD .
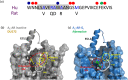
Conclusions and implications: These results demonstrate the power of the NanoBRET ligand-binding approach to study the effect of allosteric ligands on the binding of fluorescent agonists to the adenosine A1 -receptor in intact living cells. Furthermore, our studies suggest that VCP171 and PD 81,723 may switch a proportion of A1 -receptors to an active agonist conformation (R*).
© 2019 The Authors. British Journal of Pharmacology published by John Wiley & Sons Ltd on behalf of British Pharmacological Society.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Alexander, S. P. H. , Christopoulos, A. , Davenport, A. P. , Kelly, E. , Marrion, N. V. , Peters, J. A. , … CGTP Collaborators (2017). The concise guide to pharmacology 2017/18: G protein‐coupled receptors. British Journal of Pharmacology, 174(Suppl 1), S17–S129. 10.1111/bph.13878 - DOI - PMC - PubMed
-
- Aurelio, L. , Valant, C. , Flynn, B. L. , Sexton, P. M. , Christopoulos, A. , & Scammells, P. J. (2009). Allosteric modulators of the adenosine A1 receptor: Synthesis and pharmacological evaluation of 4‐substituted 2‐amino‐3‐benzoylthiophenes. Journal of Medicinal Chemistry, 52, 4543–4547. 10.1021/jm9002582 - DOI - PubMed
-
- Aurelio, L. , Valant, C. , Flynn, B. L. , Sexton, P. M. , White, J. M. , Christopoulos, A. , & Scammells, P. J. (2010). Effects of conformational restriction of 2‐amino‐3‐benzoylthiophenes on A1 adenosine receptor modulation. Journal of Medicinal Chemistry, 53, 6550–6559. 10.1021/jm1008538 - DOI - PubMed
-
- Briddon, S. J. , Middleton, R. J. , Yates, A. S. , George, M. W. , Kellam, B. , & Hill, S. J. (2004). Application of fluorescence correlation spectroscopy to the measurement of agonist binding to a G‐protein coupled receptor at the single cell level. Faraday Discussions, 26, 197–207. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

