Conditional deletion of Des1 in the mouse retina does not impair the visual cycle in cones
- PMID: 30645148
- PMCID: PMC6436658
- DOI: 10.1096/fj.201802493R
Conditional deletion of Des1 in the mouse retina does not impair the visual cycle in cones
Abstract
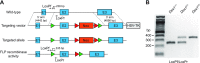
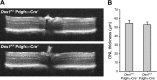
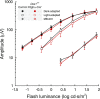
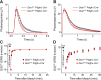
Cone photoreceptors are essential for vision under moderate to high illuminance and allow color discrimination. Their fast dark adaptation rate and resistance to saturation are believed to depend in part on an intraretinal visual cycle that supplies 11- cis-retinaldehyde to cone opsins. Candidate enzymes of this pathway have been reported, but their physiologic contribution to cone photoresponses remains unknown. Here, we evaluate the role of a candidate retinol isomerase of this pathway, sphingolipid δ4 desaturase 1 (Des1). Single-cell RNA sequencing analysis revealed Des1 expression not only in Müller glia but also throughout the retina and in the retinal pigment epithelium. We assessed cone functional dependence on Müller cell-expressed Des1 through a conditional knockout approach. Floxed Des1 mice, on a guanine nucleotide-binding protein subunit α transducin 1 knockout ( Gnat1-/-) background to allow isolated recording of cone-driven photoresponses, were bred with platelet-derived growth factor receptor α (Pdgfrα)-Cre mice to delete Des1 in Müller cells. Conditional knockout of Des1 expression, as shown by tissue-selective Des1 gene recombination and reduced Des1 catalytic activity, caused no gross changes in the retinal structure and had no effect on cone sensitivity or dark adaptation but did slightly accelerate the rate of cone phototransduction termination. These results indicate that Des1 expression in Müller cells is not required for cone visual pigment regeneration in the mouse.-Kiser, P. D., Kolesnikov, A.V., Kiser, J. Z., Dong, Z., Chaurasia, B., Wang, L., Summers, S. A., Hoang, T., Blackshaw, S., Peachey, N. S., Kefalov, V. J., Palczewski, K. Conditional deletion of Des1 in the mouse retina does not impair the visual cycle in cones.
Keywords: Müller glia; photoreceptor; retinoid cycle; sphingolipidδ(4) desaturase.
Conflict of interest statement
The authors thank David Peck and Elizabeth Bulman (both of Case Western Reserve University) for assistance with animal breeding, genotyping, and husbandry. This research was supported by funding from the U.S. Department of Veterans Affairs (IK2BX002683 to P.D.K.); a Research Career Scientist Award (to N.S.P.); the U.S. National Institutes of Health (NIH) National Eye Institute (R01EY009339 to K.P. and P.D.K., R01EY024864 to K.P., R01EY019312 to V.J.K., R01EY021126 to K.P. and V.J.K., R24EY027283 to K.P., P.D.K., V.J.K., S.B., and N.S.P., R01DK115824 to S.A.S., P30EY011373 to the Department of Ophthalmology and Visual Sciences at Case Western Reserve University, P30EY025585 to the Cleveland Clinic Cole Eye Institute, and P30EY002687 to the Department of Ophthalmology and Visual Sciences at Washington University); the American Diabetes Association (to S.A.S); the American Heart Association (to S.A.S.). The authors acknowledge departmental support from a Research to Prevent Blindness unrestricted grant. S.A.S. is a consultant and shareholder of Centaurus Therapeutics. The remaining authors declare no competing financial interests.
Figures



Similar articles
-
cis Retinol oxidation regulates photoreceptor access to the retina visual cycle and cone pigment regeneration.J Physiol. 2016 Nov 15;594(22):6753-6765. doi: 10.1113/JP272831. Epub 2016 Aug 2. J Physiol. 2016. PMID: 27385534 Free PMC article.
-
The role of retinol dehydrogenase 10 in the cone visual cycle.Sci Rep. 2017 May 24;7(1):2390. doi: 10.1038/s41598-017-02549-8. Sci Rep. 2017. PMID: 28539612 Free PMC article.
-
Non-photopic and photopic visual cycles differentially regulate immediate, early, and late phases of cone photoreceptor-mediated vision.J Biol Chem. 2020 May 8;295(19):6482-6497. doi: 10.1074/jbc.RA119.011374. Epub 2020 Apr 1. J Biol Chem. 2020. PMID: 32238432 Free PMC article.
-
The visual cycle of the cone photoreceptors of the retina.Nutr Rev. 2004 Jul;62(7 Pt 1):283-6. doi: 10.1111/j.1753-4887.2004.tb00053.x. Nutr Rev. 2004. PMID: 15384919 Review.
-
Vitamin A metabolism in rod and cone visual cycles.Annu Rev Nutr. 2012 Aug 21;32:125-45. doi: 10.1146/annurev-nutr-071811-150748. Annu Rev Nutr. 2012. PMID: 22809103 Review.
Cited by
-
Vitamin A aldehyde-taurine adduct and the visual cycle.Proc Natl Acad Sci U S A. 2020 Oct 6;117(40):24867-24875. doi: 10.1073/pnas.2005714117. Epub 2020 Sep 21. Proc Natl Acad Sci U S A. 2020. PMID: 32958638 Free PMC article.
-
Disturbed retinoid metabolism upon loss of rlbp1a impairs cone function and leads to subretinal lipid deposits and photoreceptor degeneration in the zebrafish retina.Elife. 2021 Oct 20;10:e71473. doi: 10.7554/eLife.71473. Elife. 2021. PMID: 34668483 Free PMC article.
-
Retinal pigment epithelium 65 kDa protein (RPE65): An update.Prog Retin Eye Res. 2022 May;88:101013. doi: 10.1016/j.preteyeres.2021.101013. Epub 2021 Oct 2. Prog Retin Eye Res. 2022. PMID: 34607013 Free PMC article. Review.
-
Acyl-CoA:wax alcohol acyltransferase 2 modulates the cone visual cycle in mouse retina.FASEB J. 2022 Jul;36(7):e22390. doi: 10.1096/fj.202101855RRR. FASEB J. 2022. PMID: 35665537 Free PMC article.
-
The Role of Vitamin A in Retinal Diseases.Int J Mol Sci. 2022 Jan 18;23(3):1014. doi: 10.3390/ijms23031014. Int J Mol Sci. 2022. PMID: 35162940 Free PMC article. Review.
References
-
- Hofmann K. P., Scheerer P., Hildebrand P. W., Choe H. W., Park J. H., Heck M., Ernst O. P. (2009) A G protein-coupled receptor at work: the rhodopsin model. Trends Biochem. Sci. 34, 540–552 - PubMed
-
- Kiser P. D., Zhang J., Sharma A., Angueyra J. M., Kolesnikov A. V., Badiee M., Tochtrop G. P., Kinoshita J., Peachey N. S., Li W., Kefalov V. J., Palczewski K. (2018) Retinoid isomerase inhibitors impair but do not block mammalian cone photoreceptor function. J. Gen. Physiol. 150, 571–590 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 EY011373/EY/NEI NIH HHS/United States
- P30 EY002687/EY/NEI NIH HHS/United States
- R01 EY019312/EY/NEI NIH HHS/United States
- R01 DK115824/DK/NIDDK NIH HHS/United States
- P30 EY025585/EY/NEI NIH HHS/United States
- IK2 BX002683/BX/BLRD VA/United States
- IK6 BX005233/BX/BLRD VA/United States
- R24 EY027283/EY/NEI NIH HHS/United States
- R24 EY024864/EY/NEI NIH HHS/United States
- R24 EY021126/EY/NEI NIH HHS/United States
- R01 EY025696/EY/NEI NIH HHS/United States
- R01 EY020560/EY/NEI NIH HHS/United States
- R01 EY009339/EY/NEI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

