Adding function to the genome of African Salmonella Typhimurium ST313 strain D23580
- PMID: 30645593
- PMCID: PMC6333337
- DOI: 10.1371/journal.pbio.3000059
Adding function to the genome of African Salmonella Typhimurium ST313 strain D23580
Abstract
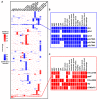
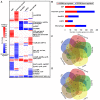
Salmonella Typhimurium sequence type (ST) 313 causes invasive nontyphoidal Salmonella (iNTS) disease in sub-Saharan Africa, targeting susceptible HIV+, malarial, or malnourished individuals. An in-depth genomic comparison between the ST313 isolate D23580 and the well-characterized ST19 isolate 4/74 that causes gastroenteritis across the globe revealed extensive synteny. To understand how the 856 nucleotide variations generated phenotypic differences, we devised a large-scale experimental approach that involved the global gene expression analysis of strains D23580 and 4/74 grown in 16 infection-relevant growth conditions. Comparison of transcriptional patterns identified virulence and metabolic genes that were differentially expressed between D23580 versus 4/74, many of which were validated by proteomics. We also uncovered the S. Typhimurium D23580 and 4/74 genes that showed expression differences during infection of murine macrophages. Our comparative transcriptomic data are presented in a new enhanced version of the Salmonella expression compendium, SalComD23580: http://bioinf.gen.tcd.ie/cgi-bin/salcom_v2.pl. We discovered that the ablation of melibiose utilization was caused by three independent SNP mutations in D23580 that are shared across ST313 lineage 2, suggesting that the ability to catabolize this carbon source has been negatively selected during ST313 evolution. The data revealed a novel, to our knowledge, plasmid maintenance system involving a plasmid-encoded CysS cysteinyl-tRNA synthetase, highlighting the power of large-scale comparative multicondition analyses to pinpoint key phenotypic differences between bacterial pathovariants.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

