The ERBB-STAT3 Axis Drives Tasmanian Devil Facial Tumor Disease
- PMID: 30645971
- PMCID: PMC6335503
- DOI: 10.1016/j.ccell.2018.11.018
The ERBB-STAT3 Axis Drives Tasmanian Devil Facial Tumor Disease
Abstract
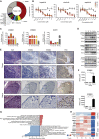
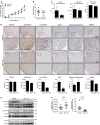
The marsupial Tasmanian devil (Sarcophilus harrisii) faces extinction due to transmissible devil facial tumor disease (DFTD). To unveil the molecular underpinnings of this transmissible cancer, we combined pharmacological screens with an integrated systems-biology characterization. Sensitivity to inhibitors of ERBB tyrosine kinases correlated with their overexpression. Proteomic and DNA methylation analyses revealed tumor-specific signatures linked to the evolutionary conserved oncogenic STAT3. ERBB inhibition blocked phosphorylation of STAT3 and arrested cancer cells. Pharmacological blockade of ERBB or STAT3 prevented tumor growth in xenograft models and restored MHC class I expression. This link between the hyperactive ERBB-STAT3 axis and major histocompatibility complex class I-mediated tumor immunosurveillance provides mechanistic insights into horizontal transmissibility and puts forward a dual chemo-immunotherapeutic strategy to save Tasmanian devils from DFTD. VIDEO ABSTRACT.
Keywords: ERBB; MHC class I; STAT3; Tasmanian devil; horizontal transmission; receptor tyrosine kinases; systems biology; transmissible cancer; tumor vulnerability; xenograft.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





Comment in
-
The Deadly Bite of STAT3.Cancer Cell. 2019 Jan 14;35(1):5-7. doi: 10.1016/j.ccell.2018.12.004. Cancer Cell. 2019. PMID: 30645976
References
-
- Ali A.M., Gomez-Biagi R.F., Rosa D.A., Lai P.S., Heaton W.L., Park J.S., Eiring A.M., Vellore N.A., de Araujo E.D. Disarming an electrophilic warhead: retaining potency in tyrosine kinase inhibitor (TKI)-resistant cml lines while circumventing pharmacokinetic liabilities. ChemMedChem. 2016;11:850–861. - PMC - PubMed
-
- Arthur-Farraj P.J., Morgan C.C., Adamowicz M., Gomez-Sanchez J.A., Fazal S.V., Beucher A., Razzaghi B., Mirsky R., Jessen K.R., Aitman T.J. Changes in the coding and non-coding transcriptome and DNA methylome that define the Schwann cell repair phenotype after nerve injury. Cell Rep. 2017;20:2719–2734. - PMC - PubMed
-
- Bae J.H., Schlessinger J. Asymmetric tyrosine kinase arrangements in activation or autophosphorylation of receptor tyrosine kinases. Mol. Cells. 2010;29:443–448. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

