Matrix association region/scaffold attachment region: the crucial player in defining the positions of chromosome breaks mediated by bile acid-induced apoptosis in nasopharyngeal epithelial cells
- PMID: 30646906
- PMCID: PMC6334432
- DOI: 10.1186/s12920-018-0465-4
Matrix association region/scaffold attachment region: the crucial player in defining the positions of chromosome breaks mediated by bile acid-induced apoptosis in nasopharyngeal epithelial cells
Abstract
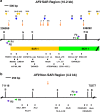
Background: It has been found that chronic rhinosinusitis (CRS) increases the risk of developing nasopharyngeal carcinoma (NPC). CRS can be caused by gastro-oesophageal reflux (GOR) that may reach nasopharynx. The major component of refluxate, bile acid (BA) has been found to be carcinogenic and genotoxic. BA-induced apoptosis has been associated with various cancers. We have previously demonstrated that BA induced apoptosis and gene cleavages in nasopharyngeal epithelial cells. Chromosomal cleavage occurs at the early stage of both apoptosis and chromosome rearrangement. It was suggested that chromosome breaks tend to cluster in the region containing matrix association region/scaffold attachment region (MAR/SAR). This study hypothesised that BA may cause chromosome breaks at MAR/SAR leading to chromosome aberrations in NPC. This study targeted the AF9 gene located at 9p22 because 9p22 is a deletion hotspot in NPC.
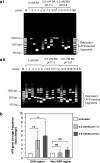
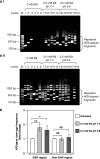
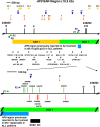
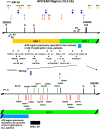
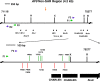
Methods: Potential MAR/SAR sites were predicted in the AF9 gene by using MAR/SAR prediction tools. Normal nasopharyngeal epithelial cells (NP69) and NPC cells (TWO4) were treated with BA at neutral and acidic pH. Inverse-PCR (IPCR) was used to identify chromosome breaks in SAR region (contains MAR/SAR) and non-SAR region (does not contain MAR/SAR). To map the chromosomal breakpoints within the AF9 SAR and non-SAR regions, DNA sequencing was performed.
Results: In the AF9 SAR region, the gene cleavage frequencies of BA-treated NP69 and TWO4 cells were significantly higher than those of untreated control. As for the AF9 non-SAR region, no significant difference in cleavage frequency was detected between untreated and BA-treated cells. A few breakpoints detected in the SAR region were mapped within the AF9 region that was previously reported to translocate with the mixed lineage leukaemia (MLL) gene in an acute lymphoblastic leukaemia (ALL) patient.
Conclusions: Our findings suggest that MAR/SAR may be involved in defining the positions of chromosomal breakages induced by BA. Our report here, for the first time, unravelled the relation of these BA-induced chromosomal breakages to the AF9 chromatin structure.
Keywords: AF9; Apoptosis; Bile acid; Chronic rhinosinusitis; Gastro-oesophageal reflux; Matrix association region/scaffold attachment region; Nasopharyngeal carcinoma.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Lo KW, Teo PM, Hui AB, To KF, Tsang YS, Chan SY, et al. High resolution allelotype of microdissected primary nasopharyngeal carcinoma. Cancer Res. 2000;60:3348–3353. - PubMed
-
- Sriamporn S, Vatanasapt V, Pisani P, Yongchaiyudha S, Rungpitarangsri V. Environmental risk factors for nasopharyngeal carcinoma: a case-control study in northeastern Thailand. Cancer Epidemiol Biomark Prev. 1992;1:345–348. - PubMed
-
- Raab-Traub N. Epstein-Barr virus and nasopharyngeal carcinoma. Semin Cancer Biol. 1992;3:297–307. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

