Machine learning algorithms for systematic review: reducing workload in a preclinical review of animal studies and reducing human screening error
- PMID: 30646959
- PMCID: PMC6334440
- DOI: 10.1186/s13643-019-0942-7
Machine learning algorithms for systematic review: reducing workload in a preclinical review of animal studies and reducing human screening error
Abstract
Background: Here, we outline a method of applying existing machine learning (ML) approaches to aid citation screening in an on-going broad and shallow systematic review of preclinical animal studies. The aim is to achieve a high-performing algorithm comparable to human screening that can reduce human resources required for carrying out this step of a systematic review.
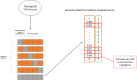
Methods: We applied ML approaches to a broad systematic review of animal models of depression at the citation screening stage. We tested two independently developed ML approaches which used different classification models and feature sets. We recorded the performance of the ML approaches on an unseen validation set of papers using sensitivity, specificity and accuracy. We aimed to achieve 95% sensitivity and to maximise specificity. The classification model providing the most accurate predictions was applied to the remaining unseen records in the dataset and will be used in the next stage of the preclinical biomedical sciences systematic review. We used a cross-validation technique to assign ML inclusion likelihood scores to the human screened records, to identify potential errors made during the human screening process (error analysis).
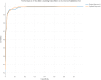
Results: ML approaches reached 98.7% sensitivity based on learning from a training set of 5749 records, with an inclusion prevalence of 13.2%. The highest level of specificity reached was 86%. Performance was assessed on an independent validation dataset. Human errors in the training and validation sets were successfully identified using the assigned inclusion likelihood from the ML model to highlight discrepancies. Training the ML algorithm on the corrected dataset improved the specificity of the algorithm without compromising sensitivity. Error analysis correction leads to a 3% improvement in sensitivity and specificity, which increases precision and accuracy of the ML algorithm.
Conclusions: This work has confirmed the performance and application of ML algorithms for screening in systematic reviews of preclinical animal studies. It has highlighted the novel use of ML algorithms to identify human error. This needs to be confirmed in other reviews with different inclusion prevalence levels, but represents a promising approach to integrating human decisions and automation in systematic review methodology.
Keywords: Analysis of human error; Automation tools; Citation screening; Machine learning; Systematic review.
Conflict of interest statement
Ethics approval
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
-
- Bornmann L, Mutz R. Growth rates of modern science: a bibliometric analysis based on the number of publications and cited references. J Assoc Inf Sci Technol. 2015;66(11):2215–2222. doi: 10.1002/asi.23329. - DOI
-
- Cohen AM, Adams CE, Davis JM, Yu C, Yu PS, Meng W, et al. Evidence-based medicine, the essential role of systematic reviews, and the need for automated text mining tools. In: Proceedings of the 1st ACM international Health Informatics Symposium: ACM; 2010. p. 376–80.
Publication types
MeSH terms
Grants and funding
- MR/N015665/1/MRC_/Medical Research Council/United Kingdom
- MR/N015665/1/WT_/Wellcome Trust/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- NC/L000970/1/NC3RS_/National Centre for the Replacement, Refinement and Reduction of Animals in Research/United Kingdom
- MR/J005037/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources

