Elucidation of the functional roles of the Q and I motifs in the human chromatin-remodeling enzyme BRG1
- PMID: 30647132
- PMCID: PMC6398141
- DOI: 10.1074/jbc.RA118.005685
Elucidation of the functional roles of the Q and I motifs in the human chromatin-remodeling enzyme BRG1
Abstract
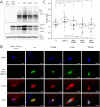
The Snf2 proteins, comprising 53 different enzymes in humans, belong to the SF2 family. Many Snf2 enzymes possess chromatin-remodeling activity, requiring a functional ATPase domain consisting of conserved motifs named Q and I-VII. These motifs form two recA-like domains, creating an ATP-binding pocket. Little is known about the function of the conserved motifs in chromatin-remodeling enzymes. Here, we characterized the function of the Q and I (Walker I) motifs in hBRG1 (SMARCA4). The motifs are in close proximity to the bound ATP, suggesting a role in nucleotide binding and/or hydrolysis. Unexpectedly, when substituting the conserved residues Gln758 (Q motif) or Lys785 (I motif) of both motifs, all variants still bound ATP and exhibited basal ATPase activity similar to that of wildtype BRG1 (wtBRG1). However, all mutants lost the nucleosome-dependent stimulation of the ATPase domain. Their chromatin-remodeling rates were impaired accordingly, but nucleosome binding was retained and still comparable with that of wtBRG1. Interestingly, a cancer-relevant substitution, L754F (Q motif), displayed defects similar to the Gln758 variant(s), arguing for a comparable loss of function. Because we excluded a mutual interference of ATP and nucleosome binding, we postulate that both motifs stimulate the ATPase and chromatin-remodeling activities upon binding of BRG1 to nucleosomes, probably via allosteric mechanisms. Furthermore, mutations of both motifs similarly affect the enzymatic functionality of BRG1 in vitro and in living cells. Of note, in BRG1-deficient H1299 cells, exogenously expressed wtBRG1, but not BRG1 Q758A and BRG1 K785R, exhibited a tumor suppressor-like function.
Keywords: ATP; ATPase; ATPase domain; BRG1; SF2 helicase family; cell cycle; cell proliferation; chromatin remodeling; nucleosome; tumor suppressor gene.
© 2019 Hoffmeister et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

