Complement Targets Newborn Retinal Ganglion Cells for Phagocytic Elimination by Microglia
- PMID: 30647151
- PMCID: PMC6507095
- DOI: 10.1523/JNEUROSCI.1854-18.2018
Complement Targets Newborn Retinal Ganglion Cells for Phagocytic Elimination by Microglia
Abstract
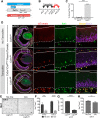
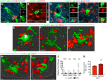
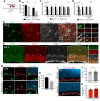
Microglia play important roles in shaping the developing CNS, and at early stages they have been proposed to regulate progenitor proliferation, differentiation, and neuronal survival. However, these studies reveal contradictory outcomes, highlighting the complexity of these cell-cell interactions. Here, we investigate microglia function during embryonic mouse retina development, where only microglia, progenitors, and neurons are present. In both sexes, we determine that microglia primarily interact with retinal neurons and find that depletion of microglia via conditional KO of the Csf1 receptor results in increased density of retinal ganglion cells (RGCs). Pharmacological inhibition of microglia also results in an increase in RGCs, with no effect on retinal progenitor proliferation, RGC genesis, or apoptosis. We show that microglia in the embryonic retina are enriched for phagocytic markers and observe engulfment of nonapoptotic Brn3-labeled RGCs. We investigate the molecular pathways that can mediate cell engulfment by microglia and find selective downregulation of complement pathway components with microglia inhibition, and further show that C1q protein marks a subset of RGCs in the embryonic retina. KO of complement receptor 3 (CR3; Itgam), which is only expressed by microglia, results in increased RGC density, similar to what we observed after depletion or inhibition of microglia. Thus, our data suggest that microglia regulate neuron elimination in the embryonic mouse retina by complement-mediated phagocytosis of non-apoptotic newborn RGCs.SIGNIFICANCE STATEMENT Microglia are emerging as active and important participants in regulating neuron number in development, during adult neurogenesis, and following stem cell therapies. However, their role in these contexts and the mechanisms involved are not fully defined. Using a well-characterized in vivo system, we provide evidence that microglia regulate neuronal elimination by complement-mediated engulfment of nonapoptotic neurons. This work provides a significant advancement of the field by defining in vivo molecular mechanisms for microglia-mediated cell elimination. Our data add to a growing body of evidence that microglia are essential for proper nervous system development. In addition, we elucidate microglia function in the developing retina, which may shed light on microglia involvement in the context of retinal injury and disease.
Keywords: complement; microglia; phagocytosis; retina; retinal ganglion cell.
Copyright © 2019 the authors 0270-6474/19/392025-16$15.00/0.
Figures





References
-
- Bosco A, Inman DM, Steele MR, Wu G, Soto I, Marsh-Armstrong N, Hubbard WC, Calkins DJ, Horner PJ, Vetter ML (2008) Reduced retina microglial activation and improved optic nerve integrity with minocycline treatment in the DBA/2J mouse model of glaucoma. Invest Ophthalmol Vis Sci 49:1437–1446. 10.1167/iovs.07-1337 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
