miRNA Expression and Interaction with Genes Involved in Susceptibility to Pristane-Induced Arthritis
- PMID: 30648118
- PMCID: PMC6311868
- DOI: 10.1155/2018/1928405
miRNA Expression and Interaction with Genes Involved in Susceptibility to Pristane-Induced Arthritis
Abstract
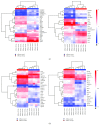
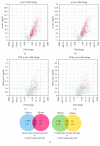
Pristane-induced arthritis (PIA) in mice is an experimental model that resembles human rheumatoid arthritis, a chronic autoimmune disease that affects joints and is characterized by synovial inflammation and articular cartilage and bone destruction. AIRmax and AIRmin mouse lines differ in their susceptibility to PIA, and linkage analysis in this model mapped arthritis severity QTLs in chromosomes 5 and 8. miRNAs are a class of small RNA molecules that have been extensively studied in the development of arthritis. We analyzed miRNA and gene expression profiles in peritoneal cells of AIRmax and AIRmin lines, in order to evaluate the genetic architecture in this model. Susceptible AIRmax mice showed higher gene (2025 vs 1043) and miRNA (240 vs 59) modulation than resistant AIRmin mice at the onset of disease symptoms. miR-132-3p/212-3p, miR-106-5p, miR-27b-3p, and miR-25-3p were among the miRNAs with the highest expression in susceptible animals, showing a negative correlation with the expression of predicted target genes (Il10, Cd69, and Sp1r1). Our study showed that global gene and miRNA expression profiles in peritoneal cells of susceptible AIRmax and resistant AIRmin lines during pristane-induced arthritis are distinct, evidencing interesting targets for further validation.
Figures




Similar articles
-
Pristane-induced arthritis loci interact with the Slc11a1 gene to determine susceptibility in mice selected for high inflammation.PLoS One. 2014 Feb 5;9(2):e88302. doi: 10.1371/journal.pone.0088302. eCollection 2014. PLoS One. 2014. PMID: 24505471 Free PMC article.
-
Slc11a1 (formerly NRAMP1) gene modulates both acute inflammatory reactions and pristane-induced arthritis in mice.Genes Immun. 2007 Jan;8(1):51-6. doi: 10.1038/sj.gene.6364358. Epub 2006 Nov 23. Genes Immun. 2007. PMID: 17122779
-
Amelioration of Experimental Autoimmune Arthritis Through Targeting of Synovial Fibroblasts by Intraarticular Delivery of MicroRNAs 140-3p and 140-5p.Arthritis Rheumatol. 2016 Feb;68(2):370-81. doi: 10.1002/art.39446. Arthritis Rheumatol. 2016. PMID: 26473405
-
miRNAs and related polymorphisms in rheumatoid arthritis susceptibility.Autoimmun Rev. 2012 Jul;11(9):636-41. doi: 10.1016/j.autrev.2011.11.004. Epub 2011 Nov 12. Autoimmun Rev. 2012. PMID: 22100329 Review.
-
MicroRNAs in rheumatoid arthritis: altered expression and diagnostic potential.Autoimmun Rev. 2015 Nov;14(11):1029-37. doi: 10.1016/j.autrev.2015.07.005. Epub 2015 Jul 8. Autoimmun Rev. 2015. PMID: 26164649 Review.
Cited by
-
Environmental Exposures and Autoimmune Diseases: Contribution of Gut Microbiome.Front Immunol. 2020 Jan 10;10:3094. doi: 10.3389/fimmu.2019.03094. eCollection 2019. Front Immunol. 2020. PMID: 31998327 Free PMC article. Review.
-
Synovial fibroblast-derived exosomal microRNA-106b suppresses chondrocyte proliferation and migration in rheumatoid arthritis via down-regulation of PDK4.J Mol Med (Berl). 2020 Mar;98(3):409-423. doi: 10.1007/s00109-020-01882-2. Epub 2020 Feb 20. J Mol Med (Berl). 2020. PMID: 32152704
-
Expression characteristics of pineal miRNAs at ovine different reproductive stages and the identification of miRNAs targeting the AANAT gene.BMC Genomics. 2021 Mar 25;22(1):217. doi: 10.1186/s12864-021-07536-y. BMC Genomics. 2021. PMID: 33765915 Free PMC article.
-
Effect of miR-132-3p on sepsis-induced acute kidney injury in mice via regulating HAVCR1/KIM-1.Am J Transl Res. 2021 Jul 15;13(7):7794-7803. eCollection 2021. Am J Transl Res. 2021. PMID: 34377256 Free PMC article.
References
-
- Picerno V., Ferro F., Adinolfi A., Valentini E., Tani C., Alunno A. One year in review: the pathogenesis of rheumatoid arthritis. Clinical and Experimental Rheumatology. 2015;33(4):551–558. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

