MSC: a metagenomic sequence classification algorithm
- PMID: 30649204
- PMCID: PMC6931357
- DOI: 10.1093/bioinformatics/bty1071
MSC: a metagenomic sequence classification algorithm
Abstract
Motivation: Metagenomics is the study of genetic materials directly sampled from natural habitats. It has the potential to reveal previously hidden diversity of microscopic life largely due to the existence of highly parallel and low-cost next-generation sequencing technology. Conventional approaches align metagenomic reads onto known reference genomes to identify microbes in the sample. Since such a collection of reference genomes is very large, the approach often needs high-end computing machines with large memory which is not often available to researchers. Alternative approaches follow an alignment-free methodology where the presence of a microbe is predicted using the information about the unique k-mers present in the microbial genomes. However, such approaches suffer from high false positives due to trading off the value of k with the computational resources. In this article, we propose a highly efficient metagenomic sequence classification (MSC) algorithm that is a hybrid of both approaches. Instead of aligning reads to the full genomes, MSC aligns reads onto a set of carefully chosen, shorter and highly discriminating model sequences built from the unique k-mers of each of the reference sequences.
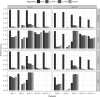
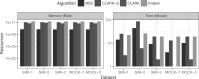
Results: Microbiome researchers are generally interested in two objectives of a taxonomic classifier: (i) to detect prevalence, i.e. the taxa present in a sample, and (ii) to estimate their relative abundances. MSC is primarily designed to detect prevalence and experimental results show that MSC is indeed a more effective and efficient algorithm compared to the other state-of-the-art algorithms in terms of accuracy, memory and runtime. Moreover, MSC outputs an approximate estimate of the abundances.
Availability and implementation: The implementations are freely available for non-commercial purposes. They can be downloaded from https://drive.google.com/open?id=1XirkAamkQ3ltWvI1W1igYQFusp9DHtVl.
© The Author(s) 2019. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures
References
-
- Buhler J., Tompa M. (2002) Finding motifs using random projections. J. Comput. Biol., 9, 225–242. - PubMed

