Conformational changes and catalytic inefficiency associated with Mot1-mediated TBP-DNA dissociation
- PMID: 30649478
- PMCID: PMC6451094
- DOI: 10.1093/nar/gky1322
Conformational changes and catalytic inefficiency associated with Mot1-mediated TBP-DNA dissociation
Abstract
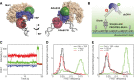
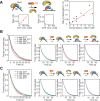
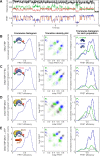
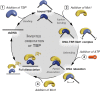
The TATA-box Binding Protein (TBP) plays a central role in regulating gene expression and is the first step in the process of pre-initiation complex (PIC) formation on promoter DNA. The lifetime of TBP at the promoter site is controlled by several cofactors including the Modifier of transcription 1 (Mot1), an essential TBP-associated ATPase. Based on ensemble measurements, Mot1 can use adenosine triphosphate (ATP) hydrolysis to displace TBP from DNA and various models for how this activity is coupled to transcriptional regulation have been proposed. However, the underlying molecular mechanism of Mot1 action is not well understood. In this work, the interaction of Mot1 with the DNA/TBP complex was investigated by single-pair Förster resonance energy transfer (spFRET). Upon Mot1 binding to the DNA/TBP complex, a transition in the DNA/TBP conformation was observed. Hydrolysis of ATP by Mot1 led to a conformational change but was not sufficient to efficiently disrupt the complex. SpFRET measurements of dual-labeled DNA suggest that Mot1's ATPase activity primes incorrectly oriented TBP for dissociation from DNA and additional Mot1 in solution is necessary for TBP unbinding. These findings provide a framework for understanding how the efficiency of Mot1's catalytic activity is tuned to establish a dynamic pool of TBP without interfering with stable and functional TBP-containing complexes.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Becker P.B., Horz W.. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 2002; 71:247–273. - PubMed
-
- Clapier C.R., Cairns B.R.. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009; 78:273–304. - PubMed
-
- Narlikar G.J., Fan H.Y., Kingston R.E.. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002; 108:475–487. - PubMed
-
- Singleton M.R., Dillingham M.S., Wigley D.B.. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007; 76:23–50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

