Biochemical and structural analyses suggest that plasminogen activators coevolved with their cognate protein substrates and inhibitors
- PMID: 30651349
- PMCID: PMC6416416
- DOI: 10.1074/jbc.RA118.005419
Biochemical and structural analyses suggest that plasminogen activators coevolved with their cognate protein substrates and inhibitors
Abstract
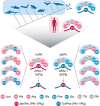
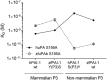
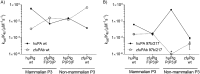
Protein sequences of members of the plasminogen activation system are present throughout the entire vertebrate phylum. This important and well-described proteolytic cascade is governed by numerous protease-substrate and protease-inhibitor interactions whose conservation is crucial to maintaining unchanged protein function throughout evolution. The pressure to preserve protein-protein interactions may lead to either co-conservation or covariation of binding interfaces. Here, we combined covariation analysis and structure-based prediction to analyze the binding interfaces of urokinase (uPA):plasminogen activator inhibitor-1 (PAI-1) and uPA:plasminogen complexes. We detected correlated variation between the S3-pocket-lining residues of uPA and the P3 residue of both PAI-1 and plasminogen. These residues are known to form numerous polar interactions in the human uPA:PAI-1 Michaelis complex. To test the effect of mutations that correlate with each other and have occurred during mammalian diversification on protein-protein interactions, we produced uPA, PAI-1, and plasminogen from human and zebrafish to represent mammalian and nonmammalian orthologs. Using single amino acid point substitutions in these proteins, we found that the binding interfaces of uPA:plasminogen and uPA:PAI-1 may have coevolved to maintain tight interactions. Moreover, we conclude that although the interaction areas between protease-substrate and protease-inhibitor are shared, the two interactions are mechanistically different. Compared with a protease cleaving its natural substrate, the interaction between a protease and its inhibitor is more complex and involves a more fine-tuned mechanism. Understanding the effects of evolution on specific protein interactions may help further pharmacological interventions of the plasminogen activation system and other proteolytic systems.
Keywords: protein evolution; serine protease; serpin; substrate specificity; surface plasmon resonance (SPR).
© 2019 Jendroszek et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
-
- Robbins K. C., Summaria L., Hsieh B., and Shah R. J. (1967) The peptide chains of human plasmin: mechanism of activation of human plasminogen to plasmin. J. Biol. Chem. 242, 2333–2342 - PubMed
-
- Robbins K. C., Bernabe P., Arzadon L., and Summaria L. (1973) NH2-terminal sequences of mammalian plasminogens and plasmin S-carboxymethyl heavy (A) and light (B) chain derivatives: a re-evaluation of the mechanism of activation of plasminogen. J. Biol. Chem. 248, 7242–7246 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

